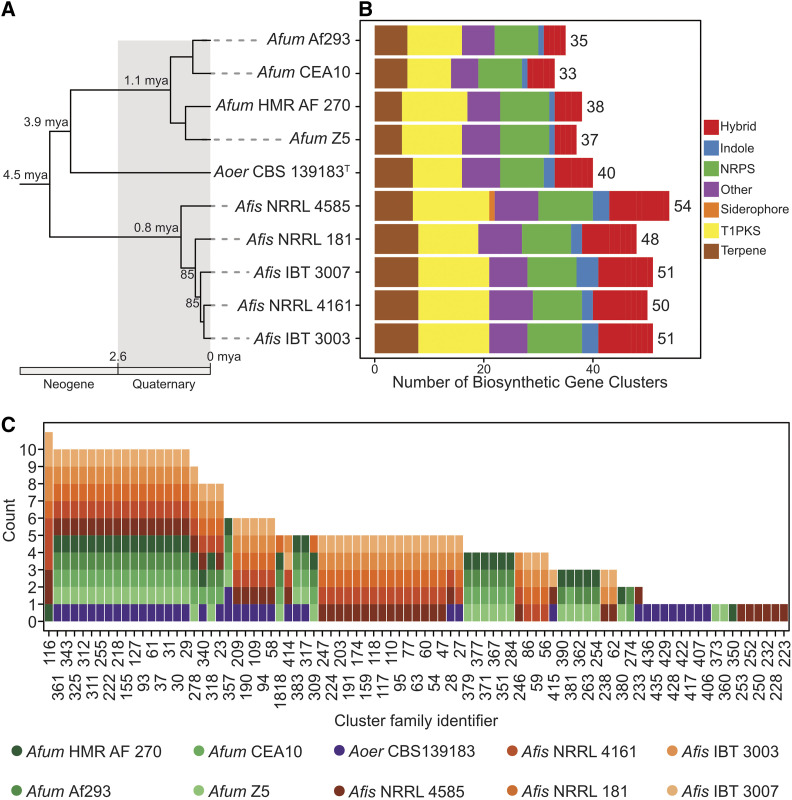
Figure 1.
Diverse genetic repertoire of biosynthetic gene clusters and extensive presence and absence polymorphisms between and within species. (A) Genome-scale phylogenomic analysis confirms A. oerlinghausenensis is the closest relative to A. fumigatus. Relaxed molecular clock analyses suggest A. fumigatus, A. oerlinghausenensis, and A. fischeri diverged from one another during the Neogene geologic period. Bipartition support is depicted for internodes that did not have full support. (B) A. fumigatus harbors the lowest number of BGCs compared to its two closest relatives. (C) Network-based clustering of BGCs into cluster families reveal extensive cluster presence and absence polymorphisms between species and strains. Cluster family identifiers are depicted on the x-axis; the number of strains represented in a cluster family are shown on the y-axis; the colors refer to a single strain from each species. Genus and species names are written using the following abbreviations: Afum, A. fumigatus; Aoer, A. oerlinghausenensis; Afis, A. fischeri. Classes of BGCs are written using the following abbreviations: NRPS, nonribosomal peptide synthetase; T1PKS, type I polyketide synthase; Hybrid, a combination of multiple BGC classes.