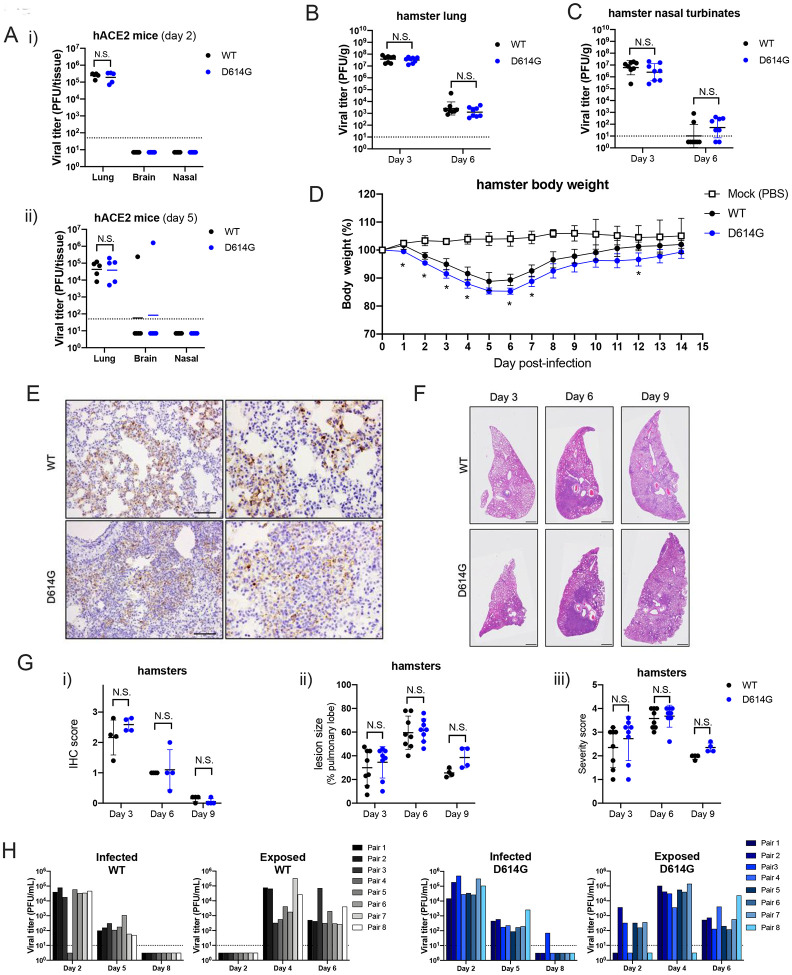
Figure 3. D614G variant exhibit similar pathogenesis but faster transmission than the WT virus in vivo.
A. Lung, brain and nasal turbinate titers of WT and D614G infected hACE2 mice were determined on day 2 (i) day 5 (ii). Each mouse was infected with 105 PFU of the virus, n= 5/group, plaque assay detection limit (1.7 log10PFU/mL) is indicated in a dash line. Viral titers of lung (B) and nasal turbinates (C) collected from SARS-CoV-2 infected hamsters at day 3 and 6. Each hamster was infected with 103 PFU of virus, plaque assay detection limit (1 log10PFU/mL) is indicated in a dash line. D. Body weight of mock-, WT- and D614G-infected hamsters, n = 4/group. E. Immunohistochemistry (IHC) staining of SARS-CoV-2 nucleocapsid protein in the representative lung tissues collected from WT- and D614G-infected hamsters, scale bar = 100 μm. F. H&E staining of representative lung tissues collected on day 3, 6, and 9 from hamsters infected with WT or D614G, scale bar : 1mm. G-i: Quantification of IHC positive cells in hamster lung tissues, following scoring system: 0, no positive cell; 1, <10%; 2, 10–50%; 3, >50% of positive cells in each lobe of lung. G-ii. The size of pulmonary lesion was determined based on the mean percentage of affected area in each section of the collected lobes from each animal. G-iii Pathological severity scores in infected hamsters, based on the percentage of inflammation area for each section of the five lobes collected from each animal following scoring system: 0, no pathological change; 1, affected area (≤10%); 2, affected area (<50%, >10%); 3, affected area (≥50%); an additional point was added when pulmonary edema and/or alveolar hemorrhage was observed. (H) Viral titers in nasal washes collected from the infected and exposed hamster pairs in WT and D614G groups; plaque assay detection limit (1 log10 PFU/mL) is indicated in a dash line. Data between the WT and D614G viruses in A, B, C, D, and G are analyzed using unpaired t-test. The number of transmitted hamsters at different timepoints are; analyzed by Fisher exact test. N.S., not significantly different, p >0.05.