FIG 6.
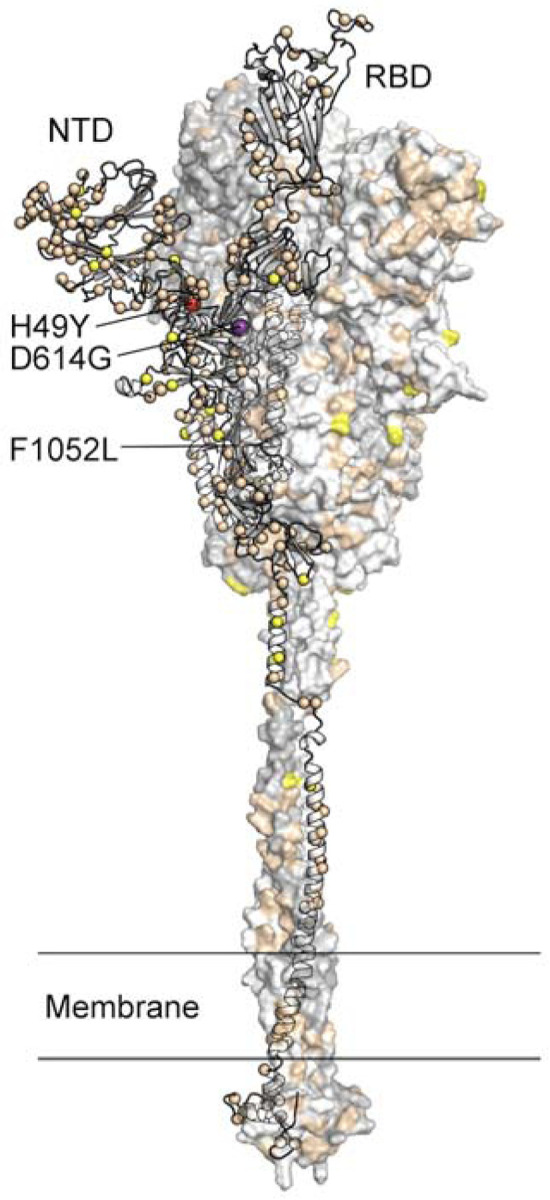
Location of amino acid substitutions mapped on the SARS-CoV-2 spike protein. Model of the SARS-CoV-2 spike protein with one protomer shown as ribbons and the other two protomers shown as a molecular surface. The Cα atom of residues found to be substituted in one or more virus isolates identified in this study is shown as a sphere on the ribbon representation. Residues found to be substituted in 1–9 isolates are colored tan, 10–99 isolates yellow, 100–999 isolates colored red (H49Y and F1052L), and >1000 isolates purple (D614G). The surface of the aminoterminal domain (NTD) that is distal to the trimeric axis has a high density of substituted residues. RBD, receptor binding domain.
