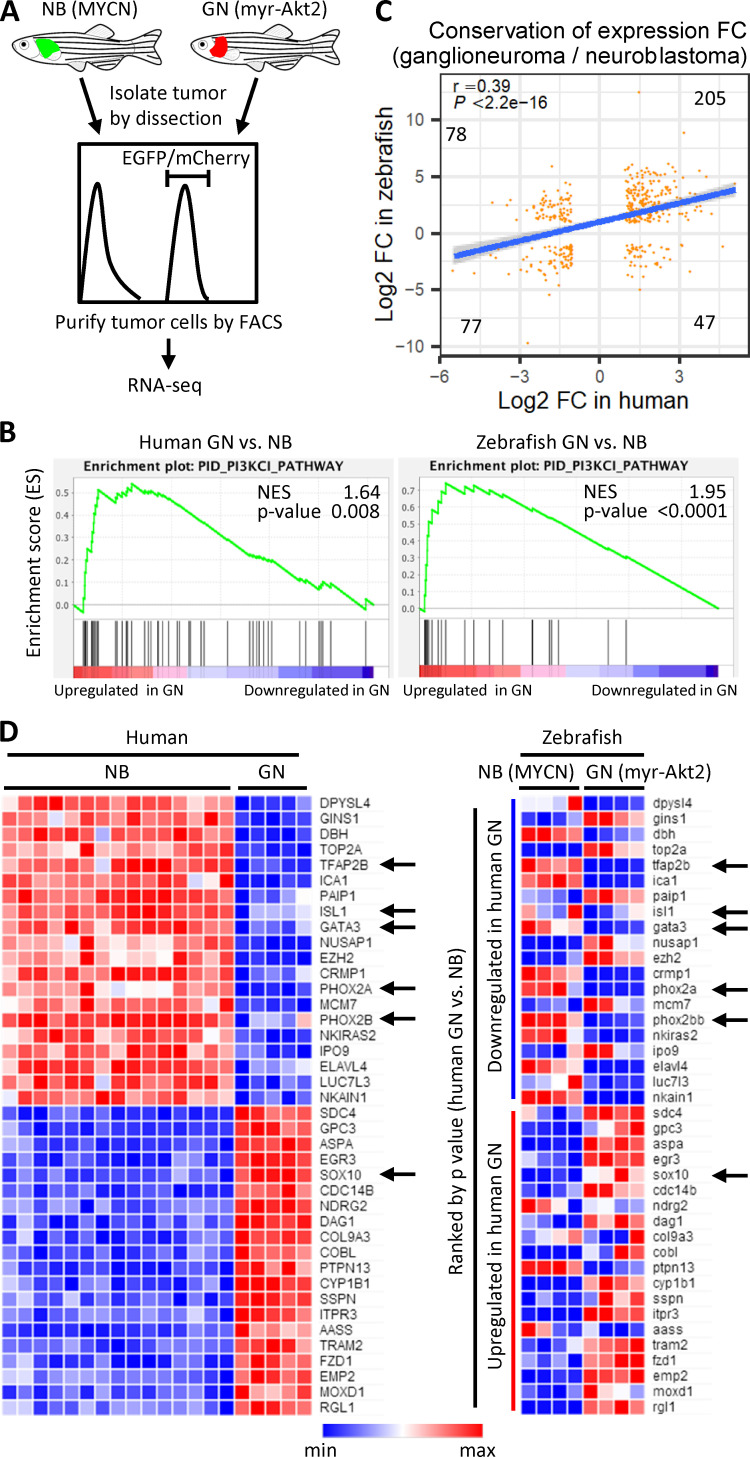
Figure 4.
Whole-transcriptome analysis of ganglioneuroma and neuroblastoma between human and zebrafish. (A) Scheme of zebrafish RNA-seq. Zebrafish neuroblastoma (NB) and ganglioneuroma (GN) cells were isolated and sorted by FACS and subjected to RNA-seq. (B) CRC to determine enrichment of a gene signature for the PI3K pathway in both human and zebrafish ganglioneuroma versus neuroblastoma samples. Genes are ranked by score and plotted along the x axis as vertical black bars. NES, normalized enrichment score. (C) Highly correlated fold change (log2 of ganglioneuroma/neuroblastoma) between human and zebrafish (Pearson’s r = 0.39, P < 2.2e-16) in gene expression of 407 Biomart one-to-one orthologues with absolute log2-fold-change >1. The linear regression line (blue) and 95% CI (surrounding shaded area) were plotted. Gene numbers within each group were indicated. (D) Heatmaps of top 20 down- or up-regulated genes (ranked by P value for human) in human and zebrafish. Arrows indicate key transcription factors in neuroblastoma and ganglioneuroma. Zebrafish RNA-seq was performed in quadruplicate for each condition.