Graphical abstract
Keywords: Docking, Molecular dynamics, DFT, HIV, YFV, Antiviral profile
Highlights
-
•
New benzimidazole analogs synthesized as antivirals against HIV-1 and yellow fever virus.
-
•
Molecular dynamics simulation studies indicated a stable ligand-protein complex of compound 3a within NNIBP of HIV-RT.
-
•
DFT analysis confirmed the stability of hydrogen bonding interaction between the TRP 229 residue of HIV-RT and compound 3a.
-
•
Molecules were tested for their anti-HIV and broad spectrum antiviral properties against different DNA and RNA viruses.
-
•
Antiviral properties and cytotoxicity determined using MTT assay.
-
•
Compound 3a showed anti-HIV activity and compound 2b showed excellent inhibition property against yellow fever virus.
Abstract
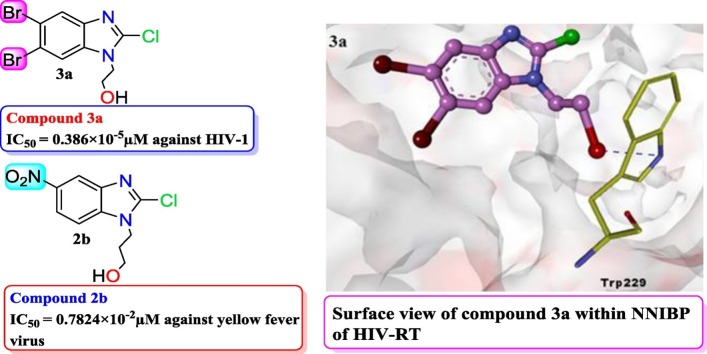
A series of alkylated benzimidazole derivatives was synthesized and screened for their anti-HIV, anti-YFV, and broad-spectrum antiviral properties. The physicochemical parameters and drug-like properties of the compounds were assessed first, and then docking studies and MD simulations on HIV-RT allosteric sites were conducted to find the possible mode of their action. DFT analysis was also performed to confirm the nature of the hydrogen bonding interaction of active compounds. The in silico studies indicated that the molecules behaved like possible NNRTIs. The nature – polar or non-polar and position of the substituent present at fifth, sixth, and N-1 positions of the benzimidazole moiety played an important role in determining the antiviral properties of the compounds. Among the various compounds, 2-(5,6-dibromo-2-chloro-1H-benzimidazol-1-yl)ethan-1-ol (3a) showed anti-HIV activity with an appreciably low IC50 value as 0.386 × 10−5μM. Similarly, compound 2b, 3-(2-chloro-5-nitro-1H-benzimidazol-1-yl) propan-1-ol, showed excellent inhibitory property against the yellow fever virus (YFV) with EC50 value as 0.7824 × 10−2μM.
1. Introduction
Most of the diseases caused by DNA and RNA viruses and the concomitant opportunistic infections represent a severe public health concern all over the world. Among viral diseases, AIDS (Acquired Immune Deficiency Syndrome) caused by human immunodeficiency virus (HIV) continues to be a deadly disease to date and stands as a major hazard to human health. Although several drugs have been discovered for the treatment of this dreadful disease, none of them were found to be efficient to kill the virus in toto, probably due to the emergence of drug resistance. To overcome this problem, several drug regimens are recommended during highly active antiretroviral therapy (HAART) used for the treatment of HIV/AIDS. As a result, improvement in adherence to medication is achieved and drug resistance has been lowered but, unfortunately, no regimen has been obtained to clean the virus out of patients completely (Tronchet and Seman, 2003; Martins et al., 2008; Zhan and Liu, 2011; Ouahrouch et al., 2016). Thus, the emergence of drug resistance warrants the discovery of new drugs that standalone or as a drug regimen focused on a single target or different targets.
In the life cycle of HIV-1, there are several drug targets but reverse transcriptase (RT) is considered to be the most critical target for anti-HIV agents because it is the enzyme responsible for viral replication through the process of reverse transcription (Kumari and Singh, 2013; Tian et al., 2014; Cichero et al., 2009a, 2009b, 2009c). HIV-RT is exposed to two major classes of inhibitors: nucleoside reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs) (Meng et al., 2014; Cichero et al., 2009a, 2009b, 2009c). NNRTIs bind to the flexible hydrophobic non-nucleoside inhibitor binding pocket (NNIBP) of RT allosteric site, 10 Å away from the active site where NRTIs bind (Cichero et al., 2009a, 2009b, 2009c). Studies on mutation in RT have confirmed that some amino acid residues (Trp229, Phe227, Leu234 and Tyr318) present in the hydrophobic region of RT, play an important role in the accurate binding of NNRTIs to the protein (RT) and make the enzyme less vulnerable to mutation (Lansdon et al., 2010; La Regina et al., 2010). So, a possible approach for designing new NNRTIs can be adopted to target this highly conserved region where the minimum possibility of mutation is expected.
Besides, the flexible and positional adaptable nature of NNRTIs plays an important role in enhancing the efficacy against a variety of drug-resistant strains of HIV-1 (Ferro et al., 2017). According to the drug design concept, a suitable inhibitor should have the efficiency to enter easily and occupy maximum space in the binding pocket to improve its potency.
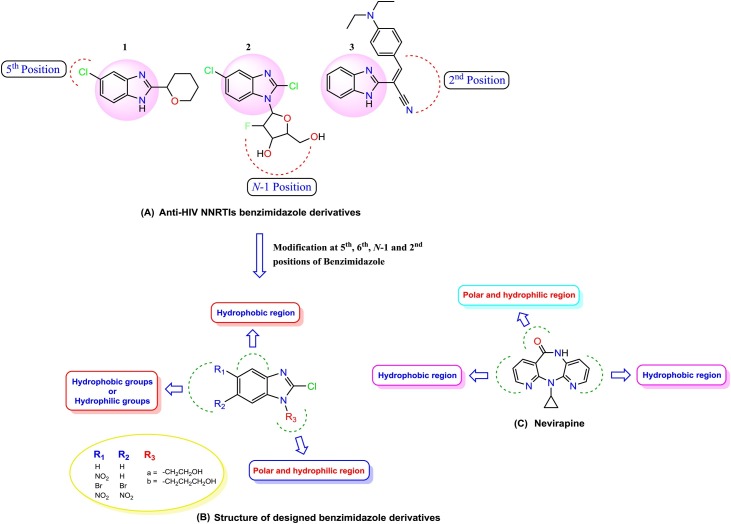
The chemistry of benzimidazole moiety is of very high importance in the field of medicinal research (Digwal et al., 2016; El-Feky et al., 2015; Evans et al., 2015; Mavrova et al., 2013; Hranjec et al., 2012; El-Gohary and Shaaban, 2014, 2015; Mahalakshmi and Chidambaranathan, 2015; Nara et al., 1989; Kus et al., 2008). Several substituted benzimidazole derivatives (Monforte et al., 2010, 2009; Monforte et al., 2008) have been reported as potent HIV-1 inhibitors (Fig. 1 ) where substitution at 5th and 6th position of benzimidazole moiety played an important role in antimicrobial/antiviral activity (Yadav and Ganguly, 2015; Akhtar et al., 2017; Francesconi et al., 2020; Cichero et al., 2017).
Fig. 1.
General structure of nevirapine and new designed benzimidazole derivatives.
We herein report the design of benzimidazole derivatives with a substituent at 5-, 6-position, and N-1 position, as possible NNRTIs against HIV-1. The synthesis of these molecules is reported in our previous work (Srivastava et al., 2018) as antibacterial agents and also given in supporting information. In the present article, we report the antiviral properties of benzimidazole derivatives.
The molecules were tested for their anti-HIV, anti-YFV and broad-spectrum antiviral properties against different DNA and RNA viruses, and the computational approaches, like molecular docking, DFT analysis, and molecular dynamics simulations were used to establish the mechanism of their mode of action.
All benzimidazole derivatives docked with HIV-RT protein interacted via hydrophilic interaction with Lys101, Lys103 or hydrophobic interaction with Tyr181, Tyr188, Phe227, Trp229 and Leu234 residues and this indicated positively towards their behaviour as NNRTIs. To compare the structural feature, docking, and biological results of these derivatives, nevirapine, a known NNRTI has been used as a reference drug.
General structures of nevirapine and N-1 alkylated benzimidazole derivatives are shown in Fig. 1.
2. Methods
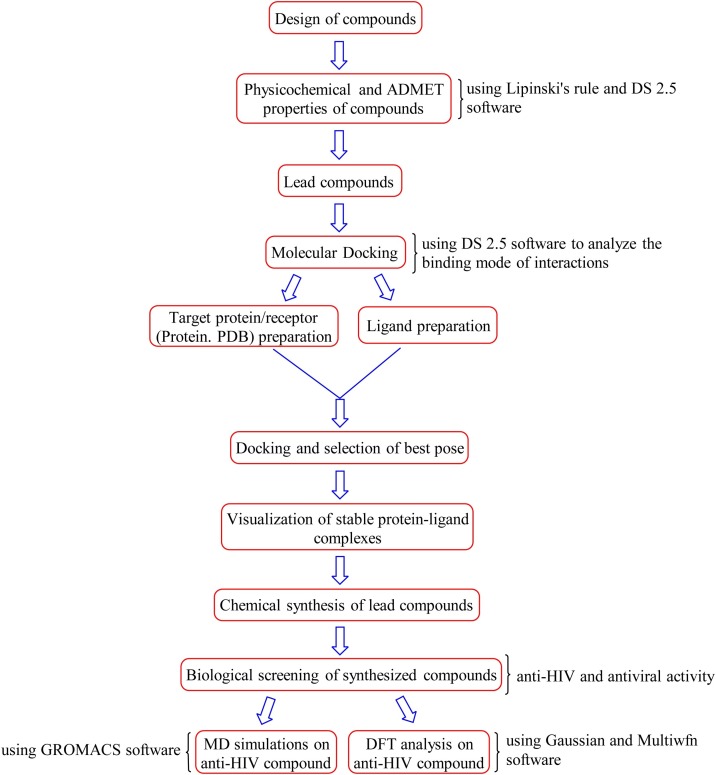
The processes involved in the development of benzimidazole derivatives as NNRTIs against HIV-1 are summarized in Fig. 2 .
Fig. 2.
Flow chart of processes involved in the development of benzimidazole derivatives.
2.1. Design, physicochemical analysis and ADMET properties of benzimidazole derivatives
Based on the Lipinski’s rule of five (Lipinski et al., 2012), twenty benzimidazole derivatives have been designed as NNRTIs against HIV-1 using Moleinspiration and Chemdraw software. After performing the in silico structure-based experiments, eight (1 ( a - b ) - 4 ( a - b )) promising molecules have been selected for further studies. The physicochemical descriptors, like molecular weight (MW), hydrogen bond donor (HBD), hydrogen bond acceptor (HBA), the logarithmic partition coefficient in octanol/water, and logP (lipophilicity) have been analyzed to evaluate the drug-like properties of the molecules (Jacomini et al., 2016). In silico ADMET (Adsorption, Distribution, Metabolism, Excretion, and Toxicity) predictions of all molecules are evaluated using DS 2.5 software to support the biological results.
2.2. Molecular docking studies
Docking studies were performed using Discovery Studio 2.5 (DS 2.5, Accelrys Ltd., UK) and 3D X-ray crystal structure of RT protein (3MEC: PDB ID) was retrieved from Protein Data Bank (www.rcsb.org). Preparation, docking, and scoring of protein and molecular dynamics (MD) simulations of protein were carried out using the published procedure (Lansdon et al., 2010; Yadav et al., 2019).
2.2.1. Preparation of the receptor
The target protein with docked ligand obtained from protein data bank was taken, extracted the ligand, and added the missing hydrogen atoms for preparing the receptor, using DS 2.5. Optimization of the position of each atom was performed using the all-atom CHARMm forcefield with Adopted Basis set Newton Raphson (ABNR) minimization algorithm until the root mean square (r.m.s.) gradient for potential energy was <0.05 kcal/mol/Å (Brooks et al., 1983). The target protein was minimized and defined as a receptor using the 'Binding Site' tool in DS 2.5. The space covered by the ligand in the receptor was defined as the binding site and an input site sphere having a radius of 5 Å was defined over the binding site. The minimized receptor created from the target protein was used for docking simulation.
2.2.2. Ligand setup
The 3D structure of each ligand was built using the built-and-edit unit of DS 2.5 and then minimized using CHARMm forcefield by the ABNR method. A conformational search of the ligands was carried out by the MD approach. The ligand was heated up to 700 K and then annealed up to 200 K and this process was repeated thirty times. After thirty cycles, the conformation of each ligand was obtained and further subjected to local energy minimization using the ABNR method. Each minimized conformation was then superimposed and the conformation with the lowest energy was selected as the most probable conformation.
2.2.3. Docking and scoring
To understand the behaviour and binding mode of benzimidazole derivatives, docking simulations were performed using DS 2.5 software. Docking is the computational prediction of the structures of ligand-protein complexes within a targeted binding site of protein receptors from the conformations of the unbound ligand and protein molecules (Kitchen et al., 2004). The DS 2.5 Ligandfit protocol, combined with a shape comparison filter and a Monte Carlo transformational search, was used for docking of the ligand with targeted protein (Venkatachalam et al., 2003). Dreiding forcefield was used to refine the docked poses by rigid body minimization of the ligand for the grid-based calculated interaction energy (Mayo et al., 1990). In the process of docking, the targeted protein receptor conformation was kept fixed. The minimization of the docked poses was carried out using the all-atom CHARMm forcefield and smart minimization method until the r.m.s. gradient for potential energy was <0.05 kcal/mol/Å. In the process of minimization, the atoms of ligand and the side chains of the residues of the receptor within 5 Å from the center of the binding site were kept flexible using simulation method (Molecular dynamics, energy minimization, and Monte Carlo simulation) (Yadav et al., 2019).
Ligand scoring is an excellent approach to assess the true conformation and the binding tightness of ligand interacted into the active site of a target protein receptor (Kitchen et al., 2004). Scoring functions, like Interaction energy, Lig_Internal_Energy, Binding energy, Dock score, and Ludi_2 and Ludi_3 values were analyzed to find out the correct pose of protein-ligand complexes. In docking simulation, Interaction energy is the energy of pairwise interaction of protein-ligand and complexes that explains the interaction of the ligand with the target protein. The internal ligand energy (LIG) of each pose of different ligands obtained from the docking simulation is the sum of the van der Walls (vdW) and electrostatic energy used for evaluating the efficiency of binding of ligands to the protein. The LUDI scores (Ludi_2 and Ludi_3) are empirical scoring functions derived from the Ludi algorithm and used to score the refined conformations of protein-ligandnd complex. The highest Ludi scores of ligand pose were selected as the best conformation. From Ludi_2 and Ludi_3 scoring functions, binding energy, and EC50 values, respectively, of compounds were predicted (Kumar et al., 2010). Docking simulations of all compounds into RT allosteric pocket generated ensembles of possible binding conformations. From these conformations, the most favourable conformation with the highest binding energy was selected for further analysis. The binding energy of ligands required for binding of ligands with the receptor in stable ligand-protein complexes (Lagos et al., 2008) was calculated by ‘Calculate Binding Energy protocol’ in DS 2.5 using the default settings (Bohm, 1994; Wang et al., 2003; Prathipati and Saxena, 2006).
2.2.4. Validation of docking protocol
The validation of the docking protocol was performed by docking the native crystallized ligand (nevirapine) and molecules under study, in the allosteric binding site of RT (Reverse transcriptase) protein receptor. The comparison of docking results with the native crystallized ligand with root mean square deviation (RMSD) < 2 Å (Clark et al., 2016; Lam et al., 2018), indicated that the used scoring function was appropriate. Docking values supported the hypothesis that experimental binding modes could be reproduced with correctness using this protocol.
2.3. Molecular dynamics simulations setup and parameters
MD simulations were carried out for the HIV-RT-compound 3a and HIV-RT-nevirapine complexes for a period of 50 ns using GROMACS (GROningen MAchine for Chemical Simulations) v5.1 molecular dynamics package (Pronk et al., 2013).The unit cell defined as a cubical box, with a minimal distance of 10 Å from the protein surface to the edges of the box, was solvated using the Simple Point Charge (SPC) water model GROMOS96 53a6 force field (Oostenbrink et al., 2004). Counter-ions were added to make every system electrically neutral at a salt concentration of 0.15 mol/L. Before the MD run, each system was subjected to energy minimization by employing the steepest descent integrator for 5000 steps with force convergence of <1000 kcal mol−1 nm−1.
Thereafter, each protein-ligand complex was equilibrated for 5 ns using canonical (NVT) and isothermal-isobaric (NPT) ensembles. During equilibration, each system was coupled with the Berendsen temperature and Parrinello-Rahman pressure controllers, respectively, to maintain temperature 300 K and pressure 1 bar. The Particle Mesh Ewald (PME) algorithm (Essmann et al., 1995) was employed to deal with the long-range Coulomb interactions with a Fourier grid spacing of 0.12 nm. The short-range van der Waals interactions were assessed by the Lennard-Jones potential with a cut-off distance of 1 nm. All bond lengths were constrained by the linear constraint solver (LINCS) method (Hess et al., 1997).
Subsequently, MD simulations were performed for 50 ns under micro-canonical ensemble by relaxing the couplings with the thermostats. In principle, the same protocol was applied to both systems. A time step of 2 fs was used and the coordinates were saved every 10 ps during the production run. For the structural analyses of every system, the resultant MD trajectories saved at the interval of 100 ps were analyzed using the built-in modules of GROMACS and visual molecular dynamics (VMD 1.9.1) (Schuler et al., 2001). The 2-D plots depicting the intrinsic dynamical stabilities captured by the root mean square deviation (RMSD), and radius of gyration (Rg) of the complexes were generated by Grace 5.1.23 program.
MD simulations were performed on HIV-1 RT with compound 3a (showing anti-HIV activity) and reference drug nevirapine for investigating the interactions and quality of the binding and affinity with RT enzyme. Three independent MD simulations were used for this study. The MD results of nevirapine were used as a control.
2.3.1. Binding and per residue interaction energy calculation
Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) was taken into consideration for free energy calculation from MD trajectory (Van Aalten et al., 1995; Wang et al., 2018). The binding free energy (ΔGbind) in a solvent medium was calculated as follows:
| ΔGbind = Gcomplex - (Gprotein + Gligand) |
Where Gcomplex is the total free energy of the substrate-protein complex, Gprotein and Gligand are the total energies of protein and substrate alone in a solvent, respectively.
The free energies for each individual Gcomplex, Gprotein and Gligand were estimated by:
| Gp = EMM + Gsolv |
where p can be protein, ligand, or complex. EMM is the average molecular mechanics potential energy in vacuum and Gsolv is the solvation free energy.
The molecular mechanics potential energy was calculated in the vacuum as follows:
| EMM = Ebonded + E non-bonded = Ebonded(Eint) + Evdw + Eelec |
Where Ebonded or (Eint) is the total bonded interaction, which includes all bonded interactions like bond, angle, dihedral and improper interactions; Enon-bonded is the total non-bonded interaction consisting of both van der Waals (Evdw) and electrostatic (Eelec) interactions. Ebonded is always taken as zero.
The solvation free energy (Gsolv) was estimated as the sum of electrostatic solvation free energy (Gpolar) and nonpolar solvation free energy (Gnon-polar) as given below:
| Gsolv = Gpolar + Gnon-polar |
Where Gpolar, the polar solvation energy, was determined using the Poisson-Boltzmann (PB) linear equation and the non-polar contribution, Gnon-polar was estimated from the solvent-accessible surface area (SASA) as per the following equation:
| Gnon-polar = γSASA + b |
Where γ (coefficient related to surface tension of the solvent) =0.02267 kJ/mol/Å2 or 0.0054 kcal/mol/Å2and b =3.849 kJ/mol or 0.916 kcal/mol
The binding free energies for all the complexes were calculated based on 5000 snapshots taken at an equal interval of time from 50 ns MD simulations.
The per-residue energy contribution was also computed to understand the contribution of individual amino acids to the total binding energy.
2.4. DFT analysis
The molecular interactions of active compound 3a with HIV-RT protein through hydrogen bonding during docking and MD simulations were further confirmed by DFT optimized structure-based theoretical calculations (Sekhar et al., 2015; Parr and Yang, 1989). The geometry optimization of complex RT:3a was carried out with the Gaussian 09 program (Frisch et al., 2009). The structure was optimized using the Density Functional Theory (DFT) method with the B3LYP/6-311 g (d,p) basis set without any solvent to get the lowest energy structure. The energy minimized structures were further confirmed by the values of vibrational frequency. To confirm the stability of the complex RT:3a through hydrogen bonding NBO, QTAIM and NCI analyses were performed using the DFT method.
2.4.1. NBO analysis
The natural bond orbital (NBO) analysis was carried out to investigate the stability of hydrogen-bonded dimers. The oxygen/nitrogen atom having lone pair acts as a donor whereas X-H (X = N, O) acts as an acceptor in the strong intermolecular charge-transfer interaction. The second-order perturbation theory was used to calculate the stabilization energies of the dimers (Galembeck et al., 2014).
2.4.2. AIM analysis
Atoms in molecules (AIM), also cited as the quantum theory of atoms in molecules (QTAIM), analysis was performed using Multiwfn program (3.3.6 version) (Lu and Chen, 2012) to generate the bond path, a bond critical point (bcp) and its corresponding energies based on the quantum observable (electron density; 〉 and the energies densities). The energies were calculated by the equation (Espinosa et al., 1998) as given below:
| E = V(bcp) / 2 |
where, V(bcp) = Potential energy at bcp generated by the Multiwfn program.
The magnitudes of electron density (〉) and the sign of Laplacian of electron density (∇2ρ) for bcp of hydrogen bonds of interest in the complex of RT:3a were calculated using AIM calculations (Lakshmipriya et al., 2015) to confirm the presence of hydrogen bonding between compound 3a and Trp229 amino acid residue of HIV-RT protein.
2.4.3. NCI analysis
The non-covalent interaction (NCI) analysis was used to detect the non-covalent interactions in real space based on the electron density and its derivatives and provided strong information about the steric repulsion, hydrogen bonding, and van der Waals interactions. It is a theoretical approach of the two-dimensional graphical plot of RDG (reduced density gradient) (s) versus sign (⌊2) × 〉 (Johnson et al., 2010).
Here, the Multiwfn program (Lu and Chen, 2012) was used to plot NCI, and VMD program (Humphrey et al., 1996) was used to plot colour filled isosurfaces for the complex RT:3a. In this analysis, the grid points were calculated and plotted for the two functions, i.e., sign (⌊2) × 〉 (function 1) and RDG (s) (function 2). The function sign (⌊2) × 〉 denoted the multiplication of electron density having the sign of second eigenvalue (⌊2) of the electron density Hessian matrix. The second function RDG was determined by the gradient of electron density (Johnson et al., 2010).
2.5. Biological screening
2.5.1. Anti-HIV assay
TZM-bl cells were used to evaluate the anti-HIV activity of N-1 alkylated benzimidazole derivatives. TZM-bl cells (4 × 104/well) were seeded in 96-well culture plate and incubated overnight at 37 °C in the presence of humidity and 5 % CO2. Further, in separate vials, HIV-1 (strain NL4.3), at a 0.05 multiplicity of infection, was treated with a variety of synthetic compounds for 1 h at 37 °C followed by the addition of pre-treated viruses in duplicate to TZM-bl cells cultured for 4 h. Nevirapine was used as a positive reference control, whereas cells without HIV were used as a negative control. After incubation, the cells were washed with cold 50 mM PBS (pH 7.4) to remove the cell-free virus, and then a fresh culture medium with or without the synthetic compounds was added to incubate cells again for 48 h. Then cells were washed two times with PBS, and lyzed with 1X lysis buffer (Promega Corporation, Madison, USA). The supernatant was analyzed for luciferase activity by luciferase assay kit (Promega Corporation). The results were expressed as percentage inhibition, calculated by taking the luminescence in an experimental set divided by the luminescence in infected cells lacking test compound multiplied by a hundred. Percent inhibition was calculated by subtracting the above value from a hundred. Furthermore, the cytotoxicity assay was also performed to calculate the selectivity index of compounds.
To evaluate cytotoxic activity of test compounds on TZM-bl cells, an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich Inc.] assay was performed. The TZM-bl cells (3 × 104 cells per well) were seeded in 96-well plates (Greiner Bio-One, GmbH, Frickenhausen, Germany) in the presence of increasing concentrations of the test compounds and incubated in an incubator at 37 °C with 5 % CO2 for 48 h. the negative control included cells treated with solvent/medium. After incubation, cell viability was measured by the addition of 20 μL MTT (5 mg/mL per well) and incubated at 37 °C for 3−5 h followed by the addition of MTT solvent (100 μL per well). The absorbance (OD) was read at 570 nm with the reference filter at 690 nm (Singh et al., 2016). Experiments were performed three times, and the percent viability was calculated using the equation given below;
2.5.2. Antiviral activity assay
The compounds were also evaluated against different viruses such as herpes simplex virus-1 (HSV-1) (KOS strain), HSV-1 thymidine kinase-deficient, acyclovir-resistant (TK− Kos, ACVr), herpes simplex virus type 2 (HSV-2) (G strains), vaccinia virus, vesicular stomatitis virus (VSV), adenoviruses-2 and coronavirus in human embryonic lung (HEL) cells; VSV, respiratory syncytial virus (RSV) and coxsackie virus B4 in human epithelial (HeLa) cells; parainfluenza-3 virus, reovirus-1, sindbis virus, Punta Toro virus, coxsackie-B4 and yellow fever virus in Vero cells (African green monkey cells); influenza virus-A (subtypes H1N1, H3N2), and influenza virus-B in MDCK cells (Madin-Darby canine kidney cells). The following reference drugs were used: brivudin, cidofovir, acyclovir, zalcitabine, ganciclovir, alovudine, UDA (Urtica dioica agglutinin), dextran sulfate (molecular weight 10,000, DS-10,000), ribavirin, mycophenolic acid, zanamivir, amantadine, and rimantadine. The antiviral assays were based on the inhibition of virus-induced cytopathicity in African green monkey kidney cells (Vero). Confluent cell cultures in microtiter 96-well plates were inoculated with 100 CCID50 of virus (1 CCID50 being the virus dose to infect 50 % of the cell cultures) and the cell cultures were incubated in the presence of varying concentrations of the test compounds. Viral-induced cytopathicity was recorded as soon as it reached completion in the control virus-infected cell cultures that were not treated with the test compounds. Antiviral activity was expressed as the EC50 or compound concentration required to reduce the virus-induced cytopathicity by 50 %.
Cytotoxicity of test compounds was expressed as the minimum cytotoxic concentration (MCC). A colorimetric 3-(4,5-dimethythiazol-2-yl)-5-(3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay system was used to calculate the 50 % cytotoxic concentration (CC50) of test compounds responsible for 50 % decrease in cell viability (Akula et al., 2017).
3. Results and discussion
3.1. Physicochemical analysis and ADMET properties of benzimidazole derivatives
All physicochemical parameters were found to be favourable for all benzimidazole derivatives (1 ( a - b ) - 4 ( a - b )). Data have been presented in Table 1 . From Table 1, it was clear that all compounds were found within the acceptable range of Lipinski’s rule of five and expected to behave as drug like molecules as nevirapine. All compounds had the potential to act as H-bond acceptors as well as donors due to the presence of −OH group, C—N and C–X dipole that helped to increase the solubility of compounds through ionization. In Table 1, the lipophilicity of all compounds indicated that compounds had no problem to passage through the cell membrane. Lower TPSA values of all compounds proved that all compounds were capable to internalize well in cells. So, all compounds were expected to possess drug-like characteristics as shown in Table 1.
Table 1.
Physiochemical data of compounds 1(a -b) - 4(a -b).
| Compound | MW | Log P | TPSA | H-A | H-D | N | Rotatable bonds | Log S | Log Kp | SAS |
|---|---|---|---|---|---|---|---|---|---|---|
| Acceptable value | < 500 | ≤ 5 | ≤ 140 | ≤ 10 | ≤ 5 | ≤ 1 | ≤ 10 | ≤ 5 | ≤ 5 | ≤ 10 |
| 1a | 196.64 | 1.86 | 38.05 | 3 | 1 | 0 | 2 | −2.06 | −4.87 | 1.72 |
| 1b | 210.66 | 2.13 | 38.05 | 3 | 1 | 0 | 3 | −2.44 | −4.68 | 1.74 |
| 2a | 241.63 | 1.80 | 83.88 | 6 | 1 | 0 | 3 | −2.32 | −4.55 | 2.15 |
| 2b | 255.66 | 2.07 | 83.88 | 6 | 1 | 0 | 4 | −2.73 | −4.15 | 2.19 |
| 3a | 354.43 | 3.39 | 38.05 | 3 | 1 | 0 | 2 | −2.57 | −4.31 | 2.00 |
| 3b | 368.46 | 3.66 | 38.05 | 3 | 1 | 0 | 3 | −2.79 | −4.08 | 2.05 |
| 4a | 286.63 | 1.69 | 129.70 | 9 | 1 | 0 | 4 | −1.69 | −4.89 | 2.35 |
| 4b | 300.66 | 1.96 | 129.70 | 9 | 1 | 0 | 5 | −2.09 | −4.77 | 2.40 |
| Nevirapine | 266.31 | 1.38 | 63.58 | 5 | 1 | 1 | 1 | −4.51 | −2.99 | 2.30 |
MW = Molecular weight, TPSA = Total polar surface area, H-A = no. of H-bond acceptors, H-D = no. of H-bond donor, N = no. of violations to Lipinski’s rule of five, Log S = Predicted aqueous solubility, Log Kp = Predicted skin permeability coefficient, SAS (Synthetic accessibility score) = From 1 (very easy) to 10 (very difficult).
To support the physicochemical properties of all benzimidazole derivatives (1 ( a - b ) - 4 ( a - b )), in silico ADMET parameters of all compounds were assessed and presented in Table 2 . Intestinal absorption was predicted to be more than 80 % for all compounds like reference drug, which was greater than the acceptable range as shown in Table 2. Compounds were considered either to behave as substrate or inhibitors of CYPs – the enzymes that play a vital role in the bioavailability of drugs. All compounds were found to be the inhibitors of CYP1A2, like the reference drug, as revealed in Table 2.
Table 2.
In Silico ADMET properties of compounds 1(a -b) - 4(a -b).
| Compound | Absorption |
Distribution |
Metabolism |
Excretion | Toxicity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water solubi lity (log mol/L) | (human) (% absorbed) | Blood brain barrier Permeabi lity (log BB) | CNS permeab ility (log PS) | CYP |
Total Clearance (log/mL/min/kg) | Oral rat chronic toxicity (LOAEL) | Hepatotoxicity (Yes/No) | |||||||
| 2D6 | 3A4 | (log mg/kg_bw/day)1A2 | 2C19 | 2C9 | 2D6 | 3A4 | ||||||||
| Substrate (Yes/No) | Inhibitor (Yes/No) | |||||||||||||
| 1a | −2.603 | 89.150 | 0.547 | −2.413 | No | No | Yes | No | No | No | No | 0.896 | 1.244 | No |
| 1b | −2.682 | 89.355 | 0.201 | −2.410 | No | No | Yes | No | No | No | No | 0.947 | 1.055 | No |
| 2a | −2.881 | 86.566 | −0.441 | −2.510 | No | No | Yes | No | No | No | No | 0.626 | 1.387 | No |
| 2b | −2.932 | 86.821 | −0.546 | −2.517 | No | No | Yes | No | No | No | No | 0.665 | 1.413 | No |
| 3a | −2.566 | 87.429 | 0.467 | −2.544 | No | No | Yes | No | No | No | No | 0.412 | 1.226 | No |
| 3b | −2.671 | 87.467 | 0.132 | −2.541 | No | No | Yes | No | No | No | No | 0.402 | 1.253 | No |
| 4a | −3.046 | 83.401 | −0.775 | −2.741 | No | No | Yes | No | No | No | No | 0.276 | 1.196 | No |
| 4b | −3.051 | 84.231 | −0.879 | −2.757 | No | No | Yes | No | No | No | No | 0.313 | 1.192 | Yes |
| Nevirapine | −3.510 | 90.421 | −0.112 | −2.881 | No | No | Yes | No | No | No | No | 0.183 | 1.015 | Yes |
Water Solubility = < -4 soluble; Intestinal absorption = Below 30 % indicates poor absorbance; Blood brain barrier Permeability = < -1considered poorly distributed to the brain; CNS (Central Nervous System) permeability = > -2 considered to penetrate the CNS; Total Clearance (logCLtot) = Lower value indicates high drug half lifetime; LOAEL (Lowest Observed Adverse Effect) = Lower value predicts minimum toxicity.
All the ADMET parameters including absorption, distribution, metabolism, excretion, and toxicity were found to be permissible for all compounds and even better in some sense than that of reference drug (Table 2).
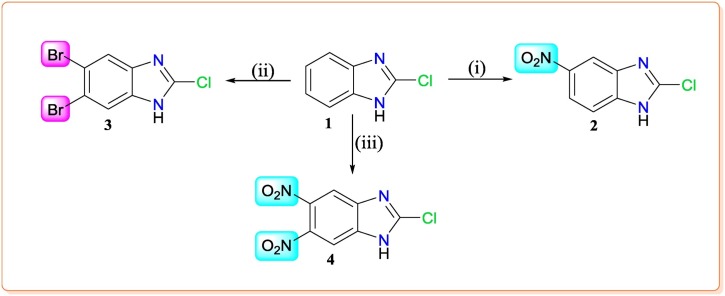
3.2. Synthesis of benzimidazole derivative (1(a - b) - 4(a - b))
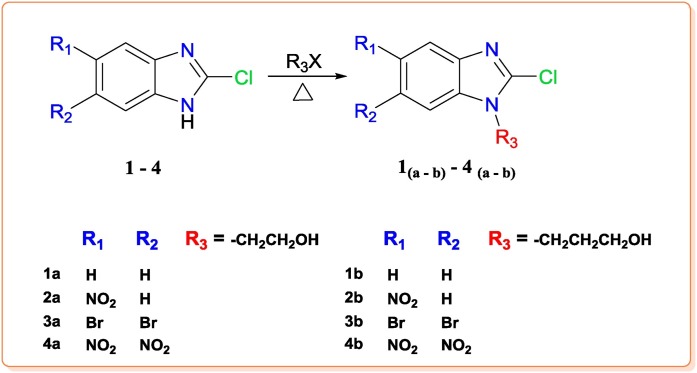
Substituted benzimidazoles 1–4 have been synthesized from 2-chloro-benzimidazole moiety. The further modifications with different substituents at 5/6 position and N-1 position on substituted benzimidazoles are clearly outlined in Scheme 1, Scheme 2 . Purification and characterization data of final benzimidazole derivatives (1 ( a - b ) - 4 ( a - b )) have been provided in supporting information.
Scheme 1.
Reagents & conditions: (i) Conc. HNO3, Conc. H2SO4, reflux for 1 h; (ii) Br2/MeOH, H2O, rt, 18 h; (iii) Conc. HNO3, Conc. H2SO4, reflux for 4–5 h.
Scheme 2.
Reagents & conditions: Alkyl halide, K2CO3, acetonitrile, Temp 40-50 °C for 3 - 4 h.
3.3. Docking analysis and scoring of N-1 alkylated benzimidazole derivatives
All benzimidazole derivatives were docked into the NNIBP (non-nucleoside inhibitor binding pocket) of the X-ray crystal structure of RT (reverse transcriptase) protein to see their mode of interaction, like hydrogen bonding and non-covalent pi - pi, pi - cation interactions that stabilize the ligand-proteinin complexes (Chen et al., 2006; Gallivan and Dougherty, 1999). The results obtained from docking simulations of all compounds with RT protein are shown in Table 3 .
Table 3.
Docking interaction of compounds 1(a -b) - 4(a - b) within NNIBP of RT protein in ligand - receptor complex.
| Compound | No. of H - B | Amino acid in H-B | Type of H-B | D(Å) | D-A | A-A | No. of π - B | Amino acid in π -B | ‘π- π’ monitor |
‘π-cation’ monitor |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bond | D(Å) | End1 | End2 | Bond | D(Å) | End1 | End2 | |||||||||
| 1a | 2 | Lys 103 | Lys 103: N - 1a: O13 1a: O13 - Lys 103: O | 2.9 2.6 | N O13 | O13 O | – | – | – | – | – | – | – | – | – | – |
| 1b | 2 | Lys 103 | Lys 103: N - 1b: O14 1b: O14 - Lys 103: O | 2.7 2.5 | N O14 | O14 O | 1 | Tyr 188 (‘π-π’) | 1b - Tyr 188 | 5.2 | 1b | Tyr 188 | – | – | – | – |
| 2a | 3 | Lys 103 Pro 236 | Lys 103: N - 2a: O16 2a: O16 - Lys 103: O 2a: O16 - Pro 236: O | 2.8 2.9 2.6 | N O16 O16 | O16 O O | – | – | – | – | – | – | – | – | – | – |
| 2b | 2 | Lys 103 Trp 229 | Lys 103: N - 2b: O12 Trp 229: N - 2b: O17 | 3.0 3.0 | N N | O12 O17 | – | – | – | – | – | – | – | – | – | – |
| 3a | 1 | Trp 229 | Trp 229: N - 3a: O13 | 3.1 | N | O13 | – | – | – | – | – | – | – | – | – | – |
| 3b | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 4a | 1 | Lys 103 | 4a: O13 - Lys 103: O | 2.6 | O13 | O | 3 | Phe 227 (‘π-cation’) Trp 229 (‘π- cation’) | – | – | – | – | Phe 227 - 4a Trp 229 - 4a Trp 229 - 4a | 6.3 5.9 6.7 | Phe 227 Trp 229 Trp 229 | 4a 4a 4a |
| 4b | 3 | Lys 101 Lys 103 Pro 236 | Lys 101: N - 4b: O13 Lys 103: N - 4b: O20 4b: O20 - Pro 236: O | 3.0 2.9 2.6 | N N O20 | O13 O20 O | – | – | – | – | – | – | – | – | – | – |
| Nevirapine (ref.) | – | – | – | – | – | – | 2 | Tyr 181 (‘π-π’) Tyr 188 (‘π-π’) | Tyr 181 - ref. Tyr 188 - ref. | 6.0 5.1 | Tyr 181 Tyr 188 | ref. ref. | – | – | – | – |
H-B = Hydrogen bond; D = Distance (Å) ; D-A = Donor Atom; A-A = Acceptor Atom; π - B = Pi bond; π-cation = Pi cation.
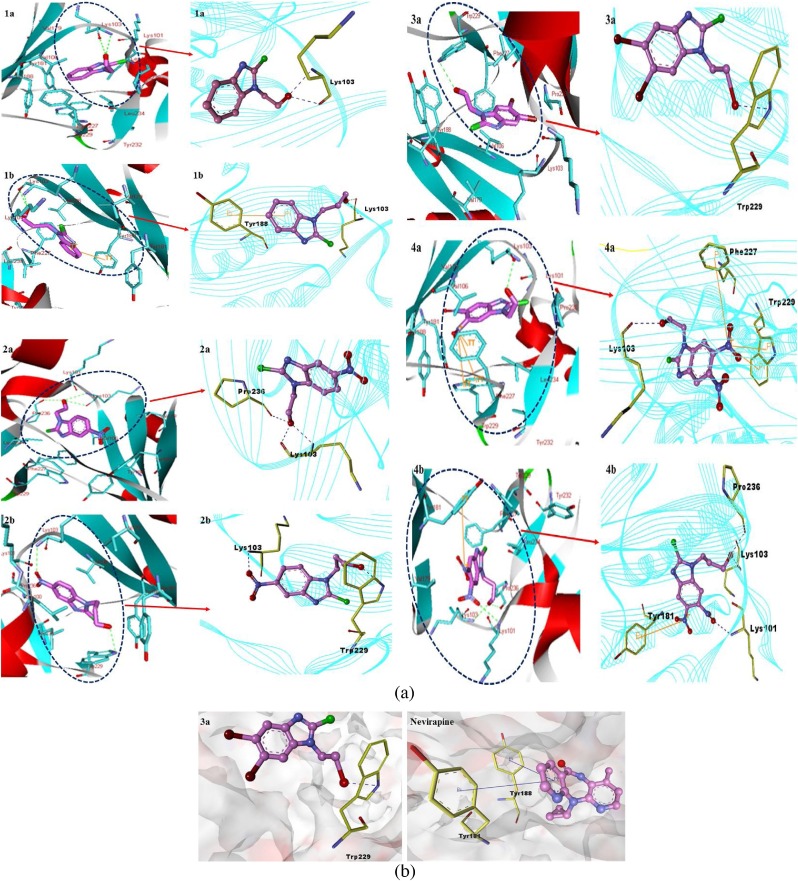
From docking simulation, it was observed that the benzimidazole ring present in all compounds was placed inside the NNIBP of HIV-RT and surrounded by the hydrophobic residues, viz. Val106, Val179, Tyr181, Tyr188, Leu234, phe227 and Trp229 and hydrophilic residues, viz. Lys101 and Lys103. The hydrophilic substituent atoms/groups present on the benzimidazole core were situated towards the outer surface of receptor site marked with the hydrophilic amino acid residues. On the other hand the hydrophobic substituent atoms/groups present on the benzimidazole core were oriented towards the hydrophobic amino acid residues present inside the hydrophobic pocket of target RT protein. Fig. 3 clearly indicated the interaction of all benzimidazole derivatives with HIV-RT proteins surrounded by the key amino acid residues, which again confirmed their binding as NNRTIs inside the NNIBP of RT protein. Results from docking simulations of compounds 1 ( a - b ) - 4 ( a - b ) revealed the most suitable orientation of all ligands. The compounds interacted with Tyr 181 and Tyr 188 amino acid residues into the hydrophobic pocket of RT protein, just like the reference drug. Additionally, some compounds showed interaction with Lys 101, Lys 103, Phe 227, Trp 229, and Pro 236 residues, which was not observed with the reference drug.
Fig. 3.
a) Docked poses with key contacts and expanded views of molecular docking interactions of compounds 1a, 1b, 2a, 2b, 3a, 4a and 4b within NNIBP of RT protein. b) Surface view of compound 3a and reference drug nevirapine within NNIBP of RT protein.
Compounds 1a and 1b interacted allosterically with Lys 103 and formed H-bonds: two H-bonds (2.9 Å and 2.6 Å) were formed by the interaction of −OH group of compound 1a and two H-bonds (2.7 Å and 2.5 Å) by the −OH group of compound 1b. Apart from this, compound 1b also interacted with Tyr 188 residue of hydrophobic pocket and showed one pi - pi interaction at a distance of 5.2 Å. Introduction of -NO2 group at the fifth position of 2-Cl-1H-benzimidazole ring of compounds 2a and 2b stabilized the protein-ligand complexes. The −OH group of compound 2a accommodated well into the binding pocket of RT protein and formed two H-bonds (2.8 Å and 2.9 Å) with a nitrogen atom and oxygen atom of Lys 103 residue, respectively, and one H-bond with the oxygen atom of Pro 236 residue at a distance of 2.6 Å. In the case of compound 2b, the oxygen atom of -NO2 group of 2-Cl-1H-benzimidazole formed an H-bond (3.0 Å) with the nitrogen atom of Lys 103 residue whereas oxygen atom of −OH group formed another H-bond (3.0 Å) with the nitrogen atom of Trp 229 residue into RT allosteric pocket. Out of compounds 3a and 3b having -Br group at the fifth and sixth position of 2-Cl-1H-benzimidazole frame, only compound 3a exhibited an H-bond (3.1 Å) through an oxygen atom of −OH group with the nitrogen atom of Trp 229 residue whereas compound 3b did not show any interaction with RT protein. One inter atomic H-bond (2.6 Å) was formed between compound 4a, having -NO2 group at fifth and sixth positions, and Lys 103. Additionally, compound 4a interacted with Phe 227 and Trp 229 residues of RT allosteric pocket and showed one pi - cation interaction (6.3 Å) and two pi - cation interactions (5.9 and 6.7 Å), respectively. Three H-bonds (3.0, 2.9 and 2.6 Å) with Lys 101, Lys 103 and Pro 236, respectively, were seen in the case of compound 4b.
Binding mode and the hydrophobic or hydrophilic contacts of benzimidazole derivatives with particular amino acid residues of HIV-RT and the reference drug nevirapine into RT allosteric pocket are shown in Fig. 3(a) and (b).
Scoring data of all molecules obtained from docking simulations are presented in Table 4 .
Table 4.
Docking scores of compounds 1(a -b) - 4(a - b) within NNIBP of RT protein in ligand - receptor complex.
| Compound | I.E. | DS | Ludi_2 | ΔG (kcal⁄ mol) | LIG | Ludi_3 | Predicted EC50 |
|---|---|---|---|---|---|---|---|
| 1a | −35.83 | 35.63 | 320 | −4.54 | −3.91 | 326 | 0.55 × 10−5 |
| 1b | −38.12 | 39.57 | 331 | −4.68 | −3.96 | 339 | 0.41 × 10−5 |
| 2a | −41.39 | 40.84 | 362 | −5.13 | −4.51 | 344 | 0.45 × 10−5 |
| 2b | −44.26 | 43.76 | 373 | −5.29 | −5.12 | 349 | 0.32 × 10−5 |
| 3a | −45.71 | 44.15 | 397 | −5.63 | −5.24 | 354 | 0.28 × 10−5 |
| 3b | −42.48 | 43.26 | 379 | −5.37 | −4.98 | 347 | 0.34 × 10−5 |
| 4a | −45.12 | 44.98 | 395 | −5.60 | −5.19 | 359 | 0.26 × 10−5 |
| 4b | −45.56 | 47.05 | 384 | −5.45 | −5.21 | 363 | 0.24 × 10−5 |
| Nevirapine | −45.79 | 40.60 | 393 | −5.57 | −5.35 | 557 | 0.27 × 10−5 |
I.E. = Interaction energy, DS = Dock score, Ludi_2 and Ludi_3 = empirical scoring functions, ΔG = Binding energy, LIG = Lig-Internal_Energy, Predicted EC50 = Predicted 50 % effective concentration of given compounds essential to reduce HIV-1 replication (μM).
From Table 4, it was observed that compounds 3a (−45.71 Kcal/mol), 4a (−45.12 Kcal/mol) and 4b (−45.56 Kcal/mol) showed almost similar interaction energy as the reference drug nevirapine (−45.79 Kcal/mol) whereas compounds 2a (−41.39 Kcal/mol), 2b (−44.26 Kcal/mol) and 3b (−42.48 Kcal/mol) showed interaction energy very close to that of the reference drug. Rest compounds showed lower interaction energy with respect to the standard drug used in molecular docking simulation.
The internal ligand energy (LIG) value of compounds 2b, 3a, 4a and 4b were somewhat comparable to that of the reference drug as shown in Table 4.
Docking results illustrated that compounds 3a (−5.63 kcal/mol) and 4a (−5.60 kcal/mol) showed higher binding free energy than the reference (−5.57 kcal/mol) whereas compounds 3b (−5.37 Kcal/mol) and 4b (−5.45 Kcal/mol) showed binding energy very close to that of the reference drug, however, the other compounds showed a little lower values of binding energy than the reference drug.
From docking simulations on test compounds, it was established that higher value of binding energy was associated with a higher affinity towards stable protein-ligand complex, which, in turn, corresponded to lower EC50 value as presented in Table 4.
Based on Dock Score, functional poses of ligand were evaluated and prioritized. It was confirmed from Table 4 that all compounds exhibited excellent docking scores ranging between 40.84 and 47.05 than the reference drug (40.60) except compounds 1a and 1b. All molecules had a similar pattern of binding into the RT allosteric site and also had a significant binding affinity with RT core, just like the reference drug. Therefore, the molecules were studied further to explore their structure-activity relationships and inhibitory activity as NNRTIs against HIV-1.
3.4. Biological screening
3.4.1. Anti-HIV assay
All N-1 alkylated benzimidazole derivatives were screened for their ability to inhibit the HIV-1 and the results obtained are summarized in Table 5 .
Table 5.
Anti-HIV-1 activity of compounds 1(a -b) - 4(a - b) evaluated in TZM-b1 Cell culture.
| Compound | Anti-HIV-1 activity |
IC50 (μg/mL) | Cytotoxicity |
CC50 (μg/mL) | SI | ||||
|---|---|---|---|---|---|---|---|---|---|
| Conc. (μg/mL) | % Inhibition |
Conc. (μg/mL) | % Viability |
||||||
| Exp - 1 | Exp - 2 | Exp - 1 | Exp - 2 | ||||||
| 1a | 10 | 2.06 | Not Tested | -- | 100 | 53.64 | Not Tested | -- | -- |
| 1 | 0.27 | -- | 10 | 90.08 | -- | -- | |||
| 1b | 10 | 4.56 | -- | 100 | 58.57 | -- | -- | ||
| 1 | 1.02 | -- | 10 | 92.63 | -- | -- | |||
| 2a | 10 | 15.55 | -- | 100 | 40.41 | -- | -- | ||
| 1 | 1.11 | -- | 10 | 103.57 | -- | -- | |||
| 2b | 10 | −1.10 | -- | 100 | 69.12 | -- | -- | ||
| 1 | −0.47 | -- | 10 | 91.23 | -- | -- | |||
| 3a | 10 | 100.47 | -- | 1.37 (0.386 × 10−5μM) | 100 | 0.96 | -- | 3.74 (1.05 × 10−5μM) | 2.73 |
| 1 | 14.91 | -- | 10 | 18.49 | 13.72 | ||||
| 10 | -- | 105.39 | 5 | -- | 49.00 | ||||
| 5 | -- | 88.54 | 1 | -- | 83.69 | ||||
| 2.5 | -- | 65.73 | -- | -- | -- | ||||
| 1.25 | -- | 33.31 | -- | -- | -- | ||||
| 3b | 10 | −4.50 | Not Tested | -- | 100 | 79.79 | Not Tested | -- | -- |
| 1 | −0.63 | -- | 10 | 101.81 | -- | -- | |||
| 4a | 10 | −6.28 | -- | 100 | 11.25 | -- | -- | ||
| 1 | −3.95 | -- | 10 | 99.91 | -- | -- | |||
| 4b | 10 | 0.11 | -- | 100 | 90.86 | -- | -- | ||
| 1 | −0.46 | -- | 10 | 92.09 | -- | -- | |||
| Nevirapine | -- | -- | -- | 1.33 × 10−9 (0.00050 × 10−5 μM) | -- | -- | -- | >2.66 (>10 μM) | >200 |
IC50 = 50 % inhibitory concentration, which is the concentration required in 50 % inhibition in HIV infection.
CC50 = 50 % cytotoxic concentration, which is the concentration required to reduce TZM-bl cell viability by 50 %.
SI = Selectivity index ratio CC50/IC50.
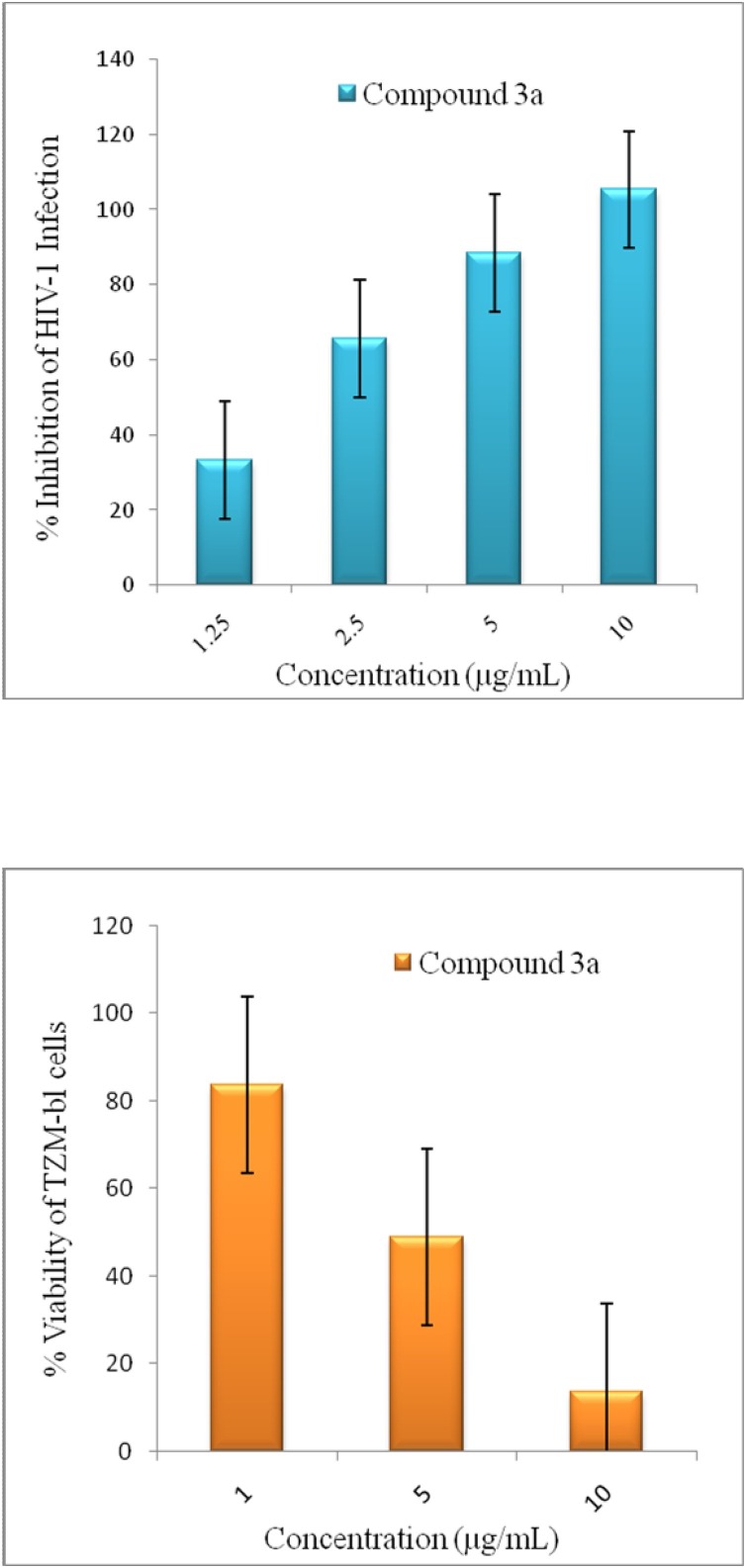
From Table 5, it was observed that amongst compounds 1 ( a - b ) - 4 ( a - b ) only compound 3a with bromo groups at 5th and 6th position and −CH2CH2OH group at N-1 position showed significant inhibition (IC50 = 0.386 × 10−5μM) against HIV-1 as compared to other compounds having -H, -NO2 substituents at 5th, 6th position and −CH2CH2CH2OH at N-1 position. However, the selectivity index (SI = 2.73) of this compound was much lower than the reference drug. All compounds showed high to moderate cytotoxicity against TZM-bl cells (Table 5). Fig. 4 showed the percentage inhibition of HIV-1 infection and cytotoxicity of compound 3a at the tested concentration.
Fig. 4.
Anti-HIV activity and cytotoxicity of compound 3a at different concentrations used in assay.
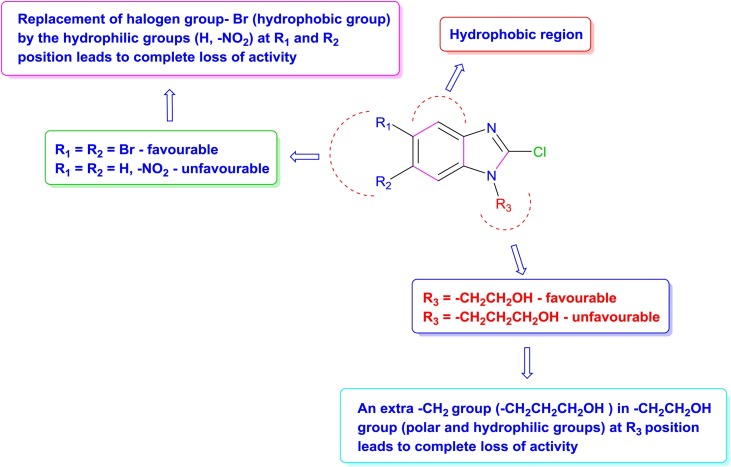
Di-bromo groups rendered higher hydrophobicity than nitro or di-nitro groups; therefore, compound 3a had higher chances for hydrophobic interactions and higher chances for H-bonding due to the presence of polar group at N-1 position with HIV RT and this could be the reason for its anti-HIV activity. However, the absence of pi-pipi interaction with Tyr181 and Tyr188 amino acid residues of the hydrophobic pocket of RT protein might be the reason for lowering its activity.
SAR studies (Fig. 5 ) confirmed the significance of the substituents at fifth, sixth and N-1 position of benzimidazole nucleus for anti-HIV activity. The structure-activity relationship study revealed that halogen groups at positions 5th, 6th and −CH2CH2OH group at position N-1 in 2-Cl-1H-benzimidazole nucleus favoured the inhibitory activity as compared to other substituents at 5th, 6th position (-H, -NO2 group) and N-1 position (−CH2CH2CH2OH group).
Fig. 5.
SAR derived from screening of test compounds against HIV-1.
3.4.2. Antiviral activity assay
The broad-spectrum antiviral activities of N-1 alkylated benzimidazole derivatives were tested against different DNA/RNA viruses and the results are given in Table 6 .
Table 6.
Antiviral activity of compounds 1 (a -b) - 4(a - b) evaluated in Vero cell culture.
| Compound | Concentration unit | Minimum cytotoxic concentrationa (MCC) | EC50b |
SIc (Yellow Fever Virus) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Para-influenza-3 virus | Reovirus-1 | Sindbis virus | Coxsackie virus B4 | Punta Toro virus | Yellow Fever virus | ||||
| 1a | μg/mL | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 1 |
| 1b | μg/mL | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 1 |
| 2a | μg/mL | ≥100 | >100 | >100 | >100 | >100 | >100 | >100 | 1 |
| 2b | μg/mL | >100 (> 0.391μM) | >100 | >100 | >100 | >100 | >100 | 6.0 (0.0234μM) | > 16.64 |
| μg/mL | ≥100 (≥ 0.391μM) | >100 | >100 | >100 | >100 | >100 | 2.0 (0.7824 × 10−2μM) | ≥ 50 | |
| 3a | μg/mL | 100 | >100 | >100 | >100 | >100 | >100 | >100 | 1 |
| 3b | μg/mL | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 1 |
| 4a | μg/mL | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 1 |
| 4b | μg/mL | ≥100 | >100 | >100 | >100 | >100 | >100 | >100 | 1 |
| DS - 10.000 | μg/mL | >100 (0.01μM) | >100 | >100 | 20 | 1.8 | 72 | 0.8 (8 × 10−5 μM) | 125 |
| Ribavirin | μM | >250 | 250 | >250 | >250 | >250 | >250 | >250 | 1 |
| Mycophenolic acid | μM | >100 | 0.6 | 6.0 | 8.9 | >100 | 14 | 0.8 | 125 |
Required to cause a microscopically detectable alteration of normal cell morphology.
Required to reduce virus-induced cytopathogenicity by 50 %.
Selectivity Index (ratio of MCC to EC50).
From Table 6, it was confirmed that all test compounds failed to show any antiviral activity against different viruses except compound 2b, which showed an inhibitory property against YFV in Vero cell cultures (Table 6). However, no activity was found in HEL, HeLa and MDCK cells.
It can be observed that compound 2b having -NO2 group at 5th position and −CH2CH2CH2OH group at N-1 position of benzimidazole nucleus had an ability to inhibit the replication of YFV at micromolar concentrations. Compound 2b showed YFV inhibition at EC50 values ranging from 0.0234μM to 0.7824 × 10−2 μM and SI values ranging from 16.64 to 50, which was higher than the reference drug ribavirin but lower than the DS-10.000 and mycophenolic acid.
3.5. Molecular dynamics simulation
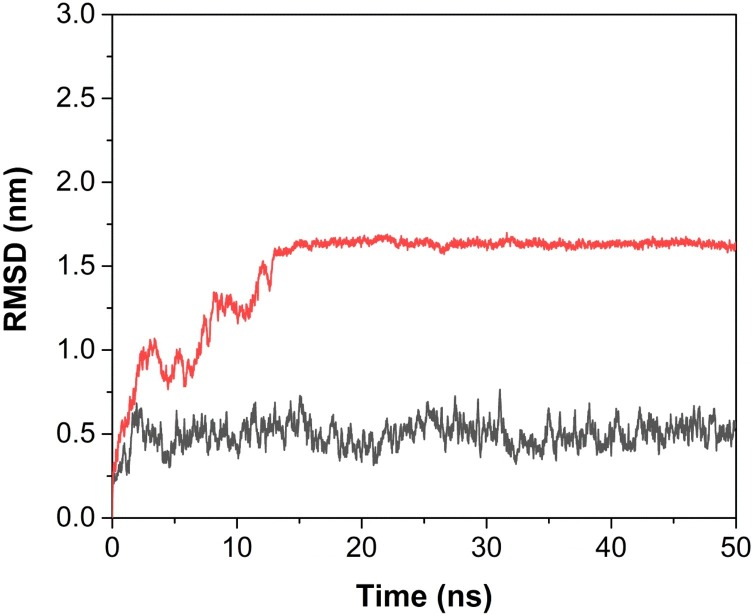
To investigate the stability of RT:nevirapine and RT:3a complexes, the RMSD values of these complexes were calculated (Fig. 6 ). Apart from assessing the equilibration, quality of the run and convergence of MD trajectories, RMSD is also useful in investigating the stability of a protein in complex.
Fig. 6.
RMSD plots of the HIV-RT complexes of nevirapine and compound 3a, (shown in black and red, respectively), as a function of time.
From Fig. 6, it was clear that there was a fluctuation in the RMSD value of RT:3a complex initially up to 1.6 nm until 15 ns, and then it got converged and remained stable throughout the 50 ns duration of MD simulation, which indicated the attainment of the stability of RT:3a complex after 15 ns.
In case of RT:nevirapine complex, fluctuation of RMSD could be seen at 20 ns, 25 ns and 35 ns, after that it remained stable till 50 ns.
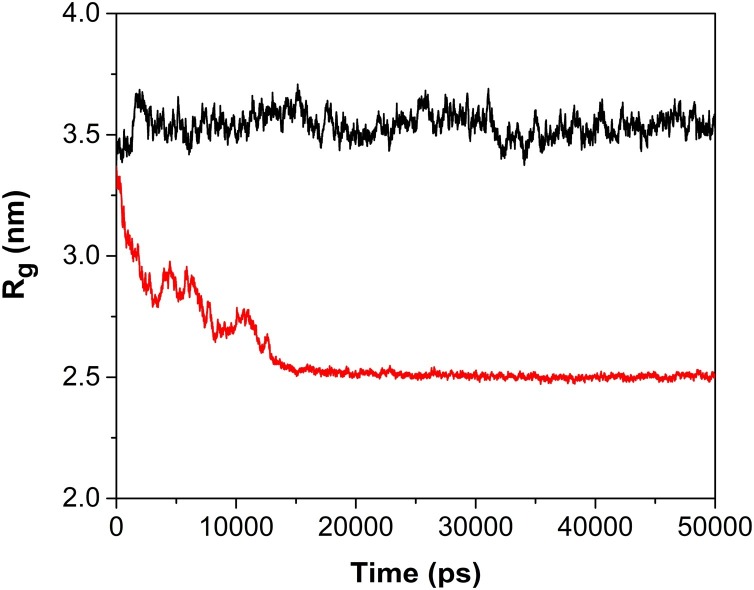
The radius of gyration (Rg) describes the level of compaction of protein. It is defined as the mass-weighted root-mean-square distance for a collection of atoms from their common center of mass. Hence, the trajectory analysis for Rg provided conformational variation, compactness and tertiary structural volume of the protein-ligand complexes. During 50 ns MD simulations, the radius of gyration for RT:3a and RT:nevirapine complexes was calculated and analyzed to see the effects of the binding mode of ligand on structural features of RT backbone (Fig. 7 ).
Fig. 7.
Rg (radius of gyration) plots of RT:nevirapine (black) and RT:compound 3a (red) complexes, as a function of time.
Like RMSD, initial fluctuation of Rg till 15 ns could be seen in the case of RT:3a complex, after that it became constant and remained stable throughout the 50 ns MD simulation. Similarly, in case of RT:nevirapine complex, fluctuation could be seen during 18–35 ns of MD simulation time period and remained stable after that.
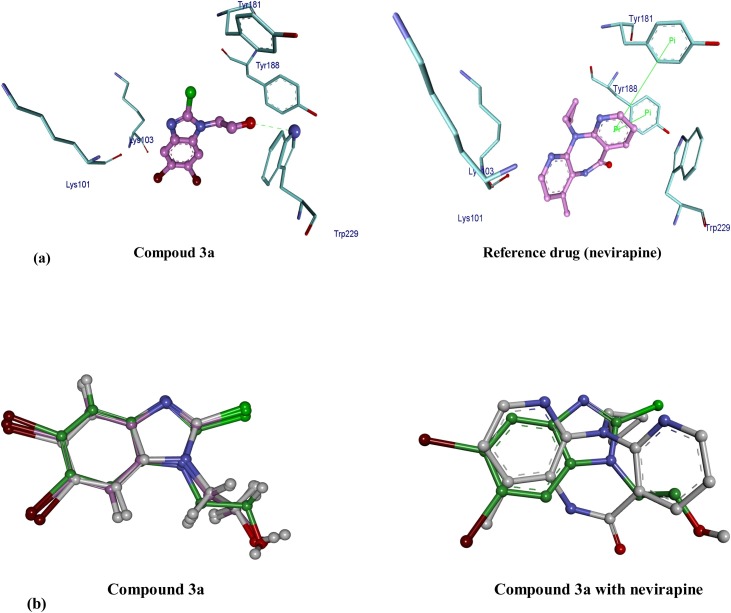
Trajectory analysis, after 50 ns, indicated that during simulations both compounds maintained their interactions with RT as observed in their docked poses (Fig. 8 ).
Fig. 8.
a) The binding mode of compound 3a and reference drug in hydrophobic pocket of RT allosteric site after 50 ns MD simulation. (Hydrogen bond and pi-pi interactions are shown by the green colour line in compound 3a and reference drug, respectively). b) Superimposition of compound 3a at different time intervals; 10, 20 and 50 ns during MD simulations and its superimposed image with nevirapine after 50 ns.
3.5.1. Binding and per residue interaction energy calculation of inhibitor:RT complexes
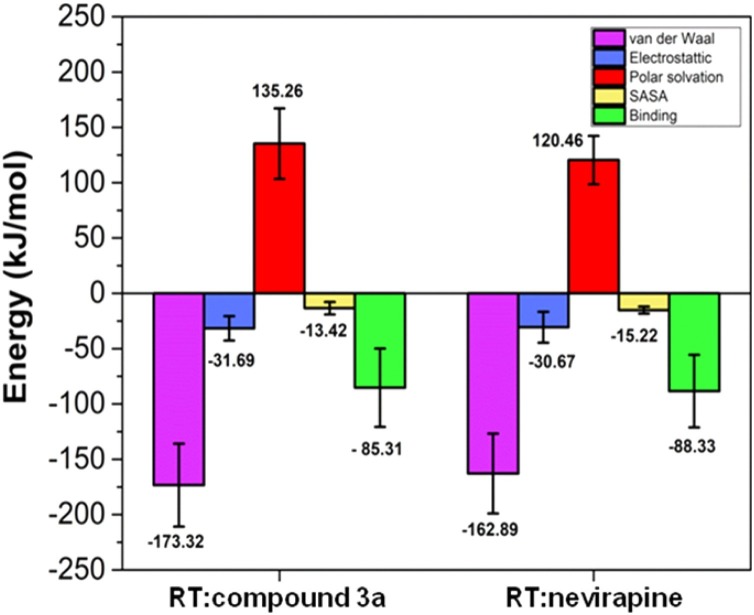
A combination of MM/PBSA and MD method was used to assess the binding free energy of RT:nevirapine and RT:3a complexes to corroborate the results. The snapshots were extracted from the 50 ns of MD simulation of each component to analyze the binding free energy of both complexes. All the energy terms of MD simulations are listed in Table 7 & Fig. 9 .
Table 7.
Molecular energy data of RT:nevirapine and RT:3a complexes.
| Energy components (kJ/mol) | RT:nevirapine | RT:3a |
|---|---|---|
| van der Waal energy (ΔGvdW) | −162.89 ± 36.07 | −173.32 ± 37.49 |
| Electrostatic energy (ΔGele) | −30.67 ± 13.97 | −31.63 ± 11.03 |
| Polar solvation energy (ΔGps) | 120.46 ± 21.84 | 135.262 ± 31.88 |
| SASA energy (ΔGsasa) | −15.24 ± 6.49 | −13.42 ± 5.94 |
| Binding energy (ΔGbinding) | −88.33 ± 32.80 | −85.31 ± 35.49 |
Fig. 9.
Error bar plot showing the van der Waals (ΔEvdW), electrostatic (ΔEelec), polar solvation, solvent accessible surface area (SASA) energies and binding free-energy (ΔGbind) in kJ/mol for all the two complexes.
In Table 7, the binding free energy (ΔGbinding) of RT:nevirapine and RT:3a complexes was -85.31 and -88.33 kJ/mol, respectively. From Table 7, it was observed that compound 3a showed almost similar calculated binding energy as that of the reference drug nevirapine. The calculated van der Waals contributions (ΔEvdw) to the binding free energy in the RT:3a (−173.32 kJ/mol) complex was more than that of RT:nevirapine complex (−162.89 kJ/mol). A sum of the calculated solvation energy (ΔEsolv), van der Waals energy (ΔEvdw) and electrostatic energy (ΔEelect) favoured the binding of compound 3a to RT allosteric site and also indicated the possibility of this molecule working as NNRTI.
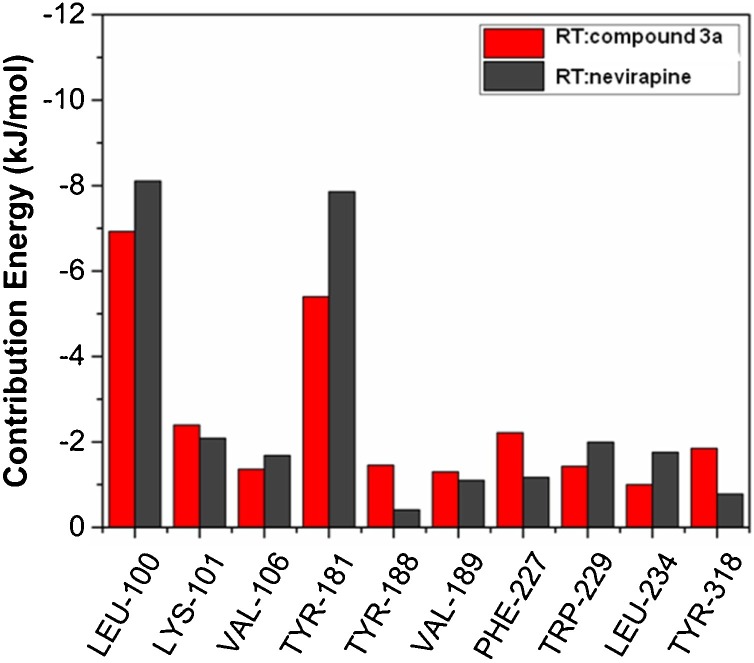
In addition to the information presented in Fig. 9 & Table 7, the contributions of individual amino acids to the binding free energy (ΔGbind) were also computed using the MM/PBSA method and presented in Fig. 10 . For clarity, only actively contributing residues towards the positive and negative binding energies are presented.
Fig. 10.
Energy contributions of individual amino acid residues to the binding free energy (ΔGbind) shown for RT:compound 3a in red, and RT:nevirapine in black color. Residues actively contributing either positive or negative to the binding energies (ΔGbind) are presented.
The per-residue interactional energy profiles revealed that Leu100, Lys101, Val106, Tyr181, Tyr188, Val189, Phe227, Trp229, Leu234 and Tyr318, actively participated in interaction and resulted in a stronger binding and stability in both the complexes. Interestingly, these residues were also interacting during the docking analysis, validating the molecular docking result.
The contribution of more than 5 kJ/mol for both RT:3a and RT:nevirapine complexes by two active residues, viz. Leu100 and Tyr181, indicated the importance of these residues in imparting stability to the complexes formed.
3.6. DFT analysis
3.6.1. NBO analysis
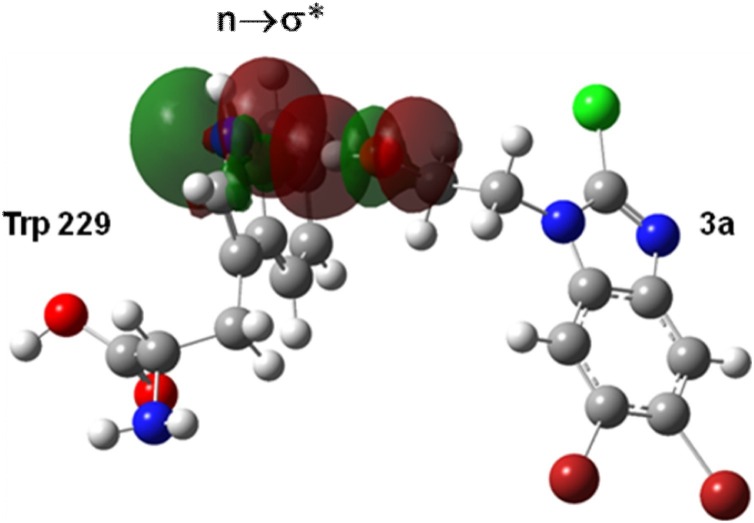
The stability of complex (RT:3a), through hydrogen bonding between compound 3a and Trp229 amino acid residue of HIV-RT protein in docking simulation, was further confirmed by NBO analysis. In NBO analysis of RT:3a complex, an important interaction from n→⌠*, i.e., (N(13)→ H(43)-O(34)) was observed. The n→⌠* interaction of a hydrogen bond involves the intermolecular delocalization (charge transfer) between a lone pair donor n and an antibonding acceptor ⌠* of an adjacent molecule, which stabilizes the structure of the protein-ligand complex.
From NBO analysis, it was found that the presence of n→⌠* interaction of hydrogen bonding was due to the transfer of lone pair electron from N-atom of Trp229 of HIV-RT to the antibonding orbital (⌠*) of O—H bond that stabilized the protein-ligand complex (RT:3a). The stabilization energy due to the process of electron transfer for the hydrogen bond is given in Table 8 and the hydrogen bonding interaction, i.e., n→⌠* (N(13)→ H(43)-O(34)) orbital overlap interaction by NBO analysis is shown in Fig. 11 .
Table 8.
Natural bond orbital analysis of orbital overlap between the Trp 229 residue of RT and compound 3a suggesting hydrogen bonding.
| Protein-ligand complex | Type of interaction | Interaction | Energy (Kcal/mol) | Software |
|---|---|---|---|---|
| Trp 229:3a | n→σ* | (N(13)→ H(43)-O(34)) | 2.94 | G09 |
Fig. 11.
NBO view showing the hydrogen bonding interaction n→⌠* (N(13)→ H(43)- O(34)) between the Trp 229 residue of RT and compound 3a.
3.6.2. AIM analysis
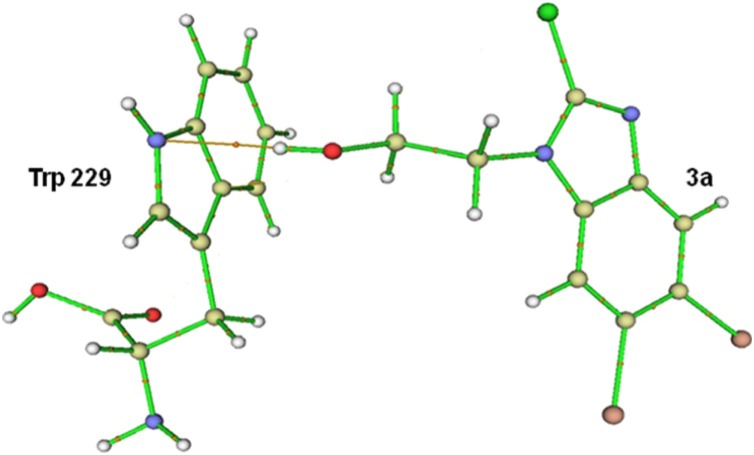
QTAIM (AIM) calculations were also carried out for hydrogen bond determination in RT:3a complex. The bcp and bond path of intermolecular hydrogen bond for the RT:3a complex is represented in Fig. 12 . The calculated values of electron density (〉) and Laplacian of electron density (∇2ρ) at each intermolecular bcp and the calculated value of hydrogen bond interaction energy (EHB) for the located bcp of hydrogen bond interest are expressed in Table 9 .
Fig. 12.
Bond critical point (bcp) and bond path between the Trp 229 residue of RT and compound 3a. (brown line denotes bond path and orange sphere between them correspond to bcp).
Table 9.
Parameters of the bcp observed between the Trp 229 residue of RT and compound 3a: distance, hydrogen bond interaction energy, electron density and the Laplacian of electron density.
| Protein-ligand complex | Interaction | Distance (Å) | Energy of HBI (Kcal/mol) | Electron density (ρ) (a.u.) | Laplacian of electron density (∇2ρ) |
|---|---|---|---|---|---|
| Trp 229:3a | (N(13)….H(43)-O(34)) | 2.0916 | 4.2031 | 0.0198 | 0.0765 |
The AIMS analysis revealed the electron density value well in the expected range (0.0102−0.0642 a.u.) for a hydrogen bond (Shahi and Arunan, 2014). The positive value of Laplacian of electron density at bcp showed that the type of interaction between compound 3a and Trp229 amino acid residue of HIV-RT was hydrogen bonding, which stabilized the formation of a protein-ligand complex.
3.6.3. NCI analysis
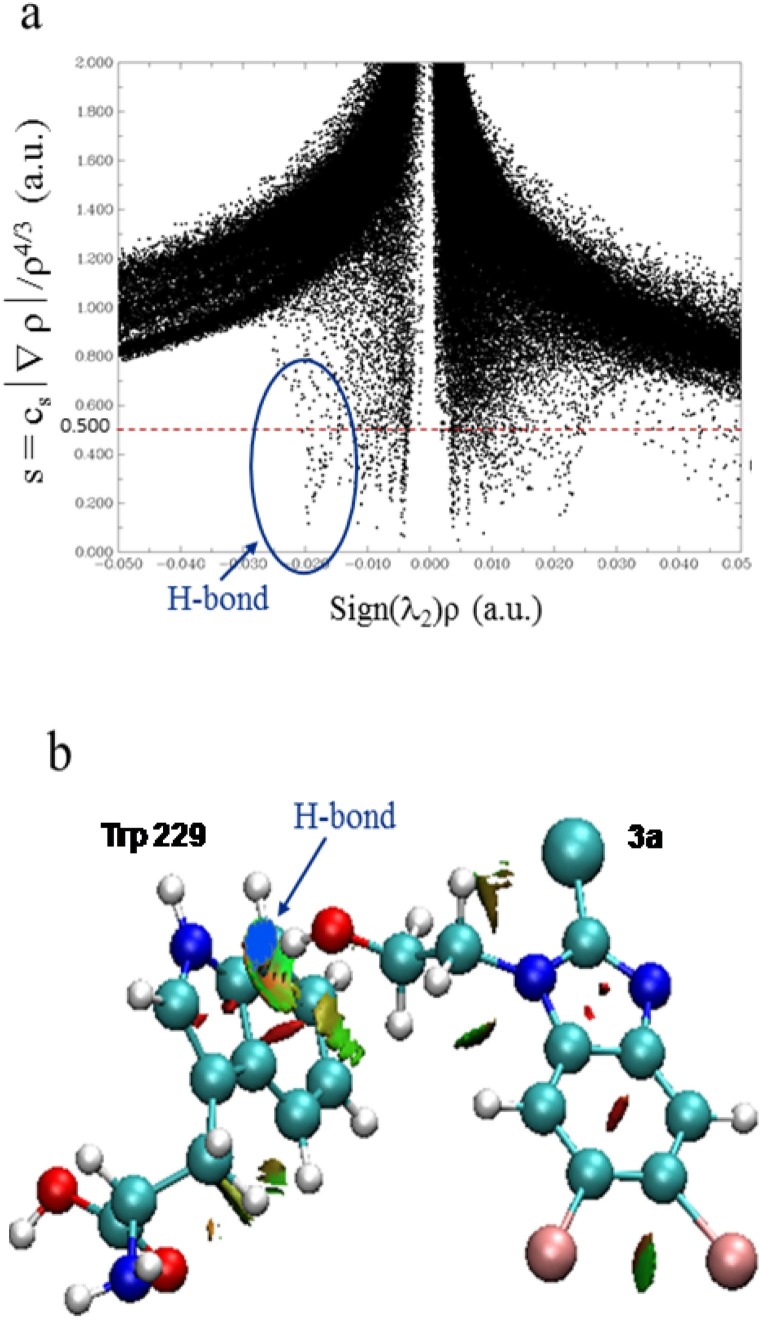
The NCI analysis was performed to affirm the presence of the hydrogen bond in the complex RT:3a. The functions carrying information about the presence of hydrogen bonding, the plot of RDG (s) versus sign (⌊2) × 〉, and the coloured gradient isosurfaces for the complex RT:3a are represented in Fig. 13 (a) and (b), respectively.
Fig. 13.
a)Plot of reduced density gradient (s) versus the electron density multiplied by the sign of the second Hessian eigen value for Trp 229 residue of RT and compound 3a complex. b)Reduced density gradient (RDG)-based NCI isosurfaces between the Trp 229 residue of RT and compound 3a. (The blue isosurfaces unambiguously characterize the short contacts with bcps as hydrogen bond interactions).
The one spike or negative peak with sign (⌊2) × 〉 ≈ −0.020 on the left-hand side in Fig. 10(a) represented one hydrogen bond, i.e., (N→ H-O) in the complex RT:3a and it can be seen in Fig. 10(b) as isosurface of blue colour. In Fig. 10(b), the red and green colour isosurfaces showed the steric interactions and van der walls interactions, respectively.
4. Conclusion
A series of N-1 alkylated benzimidazole derivatives 1 ( a - b ) - 4 ( a - b ) bearing H/NO2/Br groups at fifth and sixth position and hydroxyl alkyl groups at N-1 position was synthesized based on in silico studies. Molecular docking results indicated that binding mode of these compounds to HIV-RT allosteric site was similar to that of the reference drug, and hence, these compounds were supposed to behave as NNRTIs on the pattern of nevirapine. The compounds were screened against HIV-1 in TZM-bl cells but the expected inhibitory activity was not observed. Only the compound 3a showed anti-HIV activity with the IC50 value of 0.386 × 10−5μM but the lower SI value of this compound marred its importance. MD simulations results and DFT analysis indicated that compound 3a formed a stable complex with HIV RT, like the reference drug nevirapine. These results provided new insights to modify benzimidazole derivatives for further development of potential anti-HIV agents.
The broad-spectrum antiviral screening in Vero cell cultures revealed that compound 2b had excellent inhibition properties against the yellow fever virus with EC50 value 0.7824 × 10−2μM, equivalent to an SI value of 50.
The results found in the present study paved the way for further improvement in the antiviral property of the molecules through computational simulations.
Author contribution statement
Ritika Srivastava synthesized and purified all the compounds along with drafted the manuscript. Sunil K Gupta, Farha Naaz and Vishal K Singh did computational study (Docking and DFT analysis) of the compounds. Parth Sarthi Sen Gupta and Malay Kumar Rana performed MD simulations. Madhu Yadav and Anuradha Singh edited the manuscript. Satish K Gupta performed anti-HIV screening of all compounds. Dominique Schols performed antiviral screening. Ramendra K Singh supervised this work.
Declaration of Competing Interest
The authors have no conflict of interest.
Acknowledgements
The author Ritika Srivastava acknowledges financial support in the form of Research Fellowship from the UGC grant, University of Allahabad, Allahabad. Parth Sarthi Sen Gupta sincerely thanks IISER Berhampur, Odisha for postdoctoral fellowship.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.compbiolchem.2020.107400.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Akhtar W., Khan M.F., Verma G., Shaquiquzzaman M., Rizvi M.A., Mehdi S.H., Akhter M., Alam M.M. Therapeutic evolution of benzimidazole derivatives in the last quinquennial period. Eur. J. Med. Chem. 2017;126:705–753. doi: 10.1016/j.ejmech.2016.12.010. [DOI] [PubMed] [Google Scholar]
- Akula H.K., Kokatla H., Andrei G., Snoeck R., Schols D., Balzarini J., Lakshman M.K. Facile functionalization at the C4 position of pyrimidine nucleosides via amide group activation with (benzitriazol-1-yloxy)tris(dimethyamino)phosphonium hexafluorophosphate (BOP) and biological evaluation of the products. Org. Biomol. Chem. 2017;15(5):1130–1139. doi: 10.1039/C6OB02334G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm H.J. The development of a simple empirical scoring function to estimate the binding constant for a protein- ligand complex of known three-dimensional structure. J. Comput. Aided Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- Chen H., Lyne P.D., Giordanetto F., Lovell T., Li J. On evaluating molecular-docking methods for pose prediction and enrichment factors. J. Chem. Inf. Model. 2006;46:401–415. doi: 10.1021/ci0503255. [DOI] [PubMed] [Google Scholar]
- Cichero E., Cesarini S., Spallarossa A., Mosti L., Fossa P. Acylthiocarbamates as non nucleoside HIV-1 reverse transcriptase inhibitors: docking studies and ligand-based CoMFA and CoMSIA analyses. J. Mol. Model. 2009;15:871–884. doi: 10.1007/s00894-008-0441-6. [DOI] [PubMed] [Google Scholar]
- Cichero E., Cesarini S., Spallarossa A., Mosti L., Fossa P. Computational studies of the binding mode and 3D-QSAR analyses of symmetric formimidoester disulfides: a new class of non-nucleoside HIV-1 reverse transcriptase inhibitor. J. Mol. Model. 2009;15:357–367. doi: 10.1007/s00894-008-0402-0. [DOI] [PubMed] [Google Scholar]
- Cichero E., Cesarini S., Fossa P., Spallarossa A., Mosti L. Thiocarbamates as non nucleoside HIV-1 reverse transcriptase inhibitors: docking-based CoMFA and CoMSIA analyses. Eur. J. Med. Chem. 2009;44:2059–2070. doi: 10.1016/j.ejmech.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Cichero E., Tonelli M., Novelli F., Tasso B., Delogu I., Loddo R., Bruno O., Fossa P. Benzimidazole-based derivatives as privileged scaffold developed for the treatment of the RSV infection: a computational study exploring the potency and cytotoxicity profiles. J. Enzyme Inhib. Med. Chem. 2017;32(1):375–402. doi: 10.1080/14756366.2016.1256881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.J., Tiwary P., Borrelli K., Feng S., Miller E.B., Abel R., Friesner R.A., Berne B.J. Prediction of protein-ligand binding poses via a combination of induced fit docking and metadynamics simulations. J. Chem. Theory Comput. 2016;12:2990–2998. doi: 10.1021/acs.jctc.6b00201. [DOI] [PubMed] [Google Scholar]
- Digwal C.S., Yadav U., Sakla A.P., Ramya P.V.S., Aaghaz S., Kamal A. VOSO4 catalyzed highly efficient synthesis of benzimidazoles, benzothiazoles, and quinoxalines. Tetrahedron Lett. 2016;57:4012–4016. doi: 10.1016/j.tetlet.2016.06.074. [DOI] [Google Scholar]
- El-Feky S.A.H., El-Samii Z.K.A., Osman N.A., Lashine J., Kamel M.A., Thabet H.K. Synthesis, molecular docking and anti-inflammatory screening of novel quinoline incorporated pyrazole derivatives using the Pfitzinger reaction II. Bioorg. Chem. 2015;58:104–116. doi: 10.1016/j.bioorg.2014.12.003. [DOI] [PubMed] [Google Scholar]
- El-Gohary N.S., Shaaban M.I. Antimicrobial and antiquorum-sensing studies. Part 2: synthesis, antimicrobial, antiquorum-sensing and cytotoxic activities of new series of fused [1,3,4]thiadiazole and [1,3]benzothiazole derivatives. Med. Chem. Res. 2014;23:287–299. doi: 10.1007/s00044-013-0637-x. [DOI] [Google Scholar]
- El-Gohary N.S., Shaaban M.I. Antimicrobial and antiquorum-sensing studies. Part 3: synthesis and biological evaluation of new series of [1,3,4]thiadiazole and fused [1,3,4]thiadiazole derivatives. Arch. Pharm. Chem. Life Sci. 2015;348:283–297. doi: 10.1002/ardp.201400381. [DOI] [PubMed] [Google Scholar]
- Espinosa E., Molins E., Lecomte C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998;285:170–173. doi: 10.1016/S0009-2614(98)00036-0. [DOI] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- Evans C.W., Atkins C., Pathak A., Gilbert B.E., Noah J.W. Benzimidazole analogs inhibit respiratory syncytial virus G protein function. Antivir. Res. 2015;121:31–38. doi: 10.1016/j.antiviral.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro S., Buemi M.R., De Luca L., Agharbaoui F.E., Pannecouque C., Monforte A.M. Searching for novel N1-substituted benzimidazol-2-ones as non-nucleoside HIV-1 RT inhibitors. Bioorg. Med. Chem. 2017;25:3861–3870. doi: 10.1016/j.bmc.2017.05.040. [DOI] [PubMed] [Google Scholar]
- Francesconi V., Cichero E., Schenone S., Naesens L., Tonelli M. Synthesis and biological evaluation of novel (thio)semicarbazone-Based benzimidazoles as antiviral agents against human respiratory viruses. Molecules. 2020;25:1487. doi: 10.3390/molecules25071487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.J. 2009. Gaussian Inc. Wallingford, CT, USA. [Google Scholar]
- Galembeck S.E., Bickelhaupt F.M., Guerra C.F., Galembeck E. Effects of the protonation state in the interaction of an HIV-1 reverse transcriptase (RT) amino acid, Lys101, and a non nucleoside RT inhibitor, GW420867X. J. Mol. Model. 2014;20:2332. doi: 10.1007/s00894-014-2332-3. [DOI] [PubMed] [Google Scholar]
- Gallivan J.P., Dougherty D.A. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. U S A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- Hranjec M., Pavlovic G., Zamola G.K. Synthesis, crystal structure determination and antiproliferative activity of novel 2-amino-4-aryl-4,10-dihydro[1,3,5]triazino[1,2-a]benzimidazoles. J. Mol. Struct. 2012;1007:242–251. doi: 10.1016/j.molstruc.2011.10.054. [DOI] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jacomini A.P., Silva M.J.V., Silva R.G.M., Gonçalves D.S., Volpato H., Basso E.A., Paula F.R., Nakamura C.V., Sarragiotto M.H., Rosa F.A. Synthesis and evaluation against Leishmania amazonensis of novel pyrazolo[3,4-d]pyridazinone-N-acylhydrazone-(bi)thiophene hybrids. Eur. J. Med. Chem. 2016;124:340–349. doi: 10.1016/j.ejmech.2016.08.048. [DOI] [PubMed] [Google Scholar]
- Johnson E.R., Keinan S., Mori-Sanchez P., Contreras-Garcia J., Cohen A.J., Yang W. Revealing non-covalent interactions. J. Am. Chem. Soc. 2010;132:6498–6506. doi: 10.1021/ja100936w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen D.B., Decornez H., Furr J.R., Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nature Rev. Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- Kumar M., Vijayakrishnan R., Rao G.S. In silico structure-based design of a novel class of potent and selective small peptide inhibitor of Mycobacterium tuberculosis dihydrofolate reductase, a potential target for anti-TB drug discovery. Mol. Divers. 2010;14:595–604. doi: 10.1007/s11030-009-9172-6. [DOI] [PubMed] [Google Scholar]
- Kumari G., Singh R.K. Anti-HIV drug development: structural features and limitations of present day drugs and future challenges in the Successful HIV/AIDS treatment. Curr. Pharm. Des. 2013;19:1767–1783. doi: 10.2174/13816128113199990295. [DOI] [PubMed] [Google Scholar]
- Kus C., Kilcgil G.A., Ozbey S., Kaynak F.B., Kaya M., Coban T., Eke B.C. Synthesis and antioxidant properties of novel N-methyl-134-thiadiazol- 2-amine and 4-methyl-2H-124-triazole-3(4H)-thione derivatives of benzimidazole class. Bioorg. Med. Chem. 2008;16:4294–4303. doi: 10.1016/j.bmc.2008.02.077. [DOI] [PubMed] [Google Scholar]
- La Regina G., Coluccia A., Silvestri R. Looking for an active conformation of the future HIV type-1 non-nucleoside reverse transcriptase inhibitors. Antiviral. Chem. Chemother. 2010;20(6):213–237. doi: 10.3851/IMP1607. [DOI] [PubMed] [Google Scholar]
- Lagos C.F., Caballero J., Gonzalez-Nilo F.D., Pessoa-Mahana C.D., Perez-Acle T. Docking and quantitative structure-activity relationship studies for the bisphenylbenzimidazole family of non-nucleoside inhibitors of HIV-1 reverse transcriptase. Chem. Biol. Drug Des. 2008;72:360–369. doi: 10.1111/j.1747-0285.2008.00716.x. [DOI] [PubMed] [Google Scholar]
- Lakshmipriya A., Chaudhari S.R., Shahi A., Arunan E., Suryaprakash N. Three centered hydrogen bonds of the type C=O…H(N)…X–C in diphenyloxamide derivatives involving halogens and a rotating CF3 group: NMR, QTAIM, NCI and NBO studies. Phys. Chem. Chem. Phys. 2015;17:7528–7536. doi: 10.1039/C4CP05917D. [DOI] [PubMed] [Google Scholar]
- Lam P.C.‑H., Abagyan R., Totrov M. Ligand-biased ensemble receptor docking (LigBEnD): a hybrid ligand/receptor structure-based approach. J. Comput. Aided Mol. Des. 2018;32:187–198. doi: 10.1007/s10822-017-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdon E.B., Brendza K.M., Hung M., Wang R., Mukund S., Jin D., Birkus G., Kutty N., Liu X. Crystal structureof HIV-1 reverse transcriptase with etravirine (TMC 125) and relpivirine (TMC 278): implications for drug design. J. Med. Chem. 2010;53(10):4295–4299. doi: 10.1021/jm1002233. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lambardo F., Dominy B.W., Fenney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lu T., Chen F. Multiwfn: a multifunctional wave function analyzer. J. Comput. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- Mahalakshmi C.M., Chidambaranathan V. Synthesis, spectral characterization and antimicrobial studies of novel benzimidazole derivatives. Int. J. Chem. Sci. 2015;13:205–212. [Google Scholar]
- Martins S., Ramos M.J., Fernandes P.A. The current status of the NNRTI family of antiretrovirals used against HIV infection. Curr. Med. Chem. 2008;15(11):1083–1095. doi: 10.2174/092986708784221467. [DOI] [PubMed] [Google Scholar]
- Mavrova A.T., Wesselinova D., Vassilev N., Tsenov J.A. Design, synthesis and antiproliferative properties of some new 5-substituted-2-iminobenzimidazole derivatives. Eur. J. Med. Chem. 2013;63:696–701. doi: 10.1016/j.ejmech.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Mayo S.L., Olafson B.D., Goddard W.A.I.I.I. DREIDING: a generic force field for molecular simulations. J. Phys. Chem. 1990;94:8897–8909. doi: 10.1021/j100389a010. [DOI] [Google Scholar]
- Meng G., Liu Y., Zheng A., Chen F., Chen W., De Clercq E., Pannecouque C., Balzarini J. Design and synthesis of a new series of modified CH-diarylpyrimidines as drug-resistant HIV non-nucleoside reverse transcriptase inhibitors. Eur. J. Med. Chem. 2014;82:600–611. doi: 10.1016/j.ejmech.2014.05.059. [DOI] [PubMed] [Google Scholar]
- Monforte A.M., Rao A., Logoteta P., Ferro S., De Luca L., Barreca M.L., Iraci N., Maga G., De Clercq E., Pannecouque C., Chimirri A. Novel N1-substituted 1,3-dihydro-2H-benzimidazol-2-ones as potent non-nucleoside reverse transcriptase inhibitors. Bioorg. Med. Chem. 2008;16(15):7429–7435. doi: 10.1016/j.bmc.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Monforte A.M., Logoteta P., Ferro S., De Luca L., Iraci N., Maga G., De Clercq E., Pannecouque C., Chimirri A. Design, synthesis, and structure-activity relationships of 1,3-dihydrobenzimidazol-2-one analogues as anti-HIV agents. Bioorg. Med. Chem. 2009;17(16):5962–5967. doi: 10.1016/j.bmc.2009.06.068. [DOI] [PubMed] [Google Scholar]
- Monforte A.M., Logoteta P., De Luca L., Iraci N., Ferro S., Maga G., De Clercq E., Pannecouque C., Chimirri A. Novel 1,3-dihydro-benzimidazol-2-ones and their analogues as potent non-nucleoside HIV-1 reverse transcriptase inhibitors. Bioorg. Med. Chem. 2010;18(4):1702–1710. doi: 10.1016/j.bmc.2009.12.059. [DOI] [PubMed] [Google Scholar]
- Nara, N.S., Mukoh, H.K., Ikeda, M.A., 1989. 2-(4-Pyridylaminomethyl)benzimidazole derivatives having antiviral activity. US patent, 4,818,761, Apr. 4.
- Oostenbrink C., Villa A., Mark A.E., Van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- Ouahrouch A., Taourirte M., Schols D., Snoeck R., Andrei G., Engels J.W., Lazrek H.B. Design, synthesis and antiviral activity of novel ribonucleosides of 1,2,3-Triazolylbenzyl-aminophosphonates. Arch. Pharm. Chem. Life Sci. 2016;349:30–41. doi: 10.1002/ardp.201500292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr R.G., Yang W. Oxford University Press; New York: 1989. Density-Functional Theory of Atoms and Molecules. (1989) [Google Scholar]
- Prathipati P., Saxena A.K. Evaluation of binary QSAR models derived from LUDI and MOE scoring functions for structure based virtual screening. J. Chem. Inf. Model. 2006;46:39–51. doi: 10.1021/ci050120w. [DOI] [PubMed] [Google Scholar]
- Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation tool kit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler L.D., Daura X., Van Gunsteren W.F. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J. Comput. Chem. 2001;22:1205–1218. doi: 10.1002/jcc.1078. [DOI] [Google Scholar]
- Sekhar M.C., Venkatesulu A., Madhu Mohan T., Gowrisankar M. Density functional theory, natural Bond Orbital and atoms in molecule analyses on the hydrogen bonding interactions in 2-chloroaniline-carboxylic acids. Orient. J. Chem. 2015;31(2):897–906. doi: 10.13005/ojc/310233. [DOI] [Google Scholar]
- Shahi A., Arunan E. Hydrogen bonding, halogen bonding and lithium bonding: an atoms in molecules and natural bond orbital perspective towards conservation of total bond order, inter- and intra-molecular bonding. Phys. Chem. Chem. Phys. 2014;16:22935–22952. doi: 10.1039/C4CP02585G. [DOI] [PubMed] [Google Scholar]
- Singh A., Yadav M., Srivastava R., Singh N., Kaur R., Gupta S.K., Singh R.K. Design and anti-HIV activity of aryl sulphonamides as non-nucleoside reverse transcriptase inhibitors. Med. Chem. Res. 2016;25:2842–2859. doi: 10.1007/s00044-016-1707-7. [DOI] [Google Scholar]
- Srivastava R., Gupta S.K., Naaz F., Singh A., Singh V.K., Verma R., Singh N., Singh R.K. Synthesis, antibacterial activity, synergistic effect, cytotoxicity, docking and molecular dynamics of benzimidazole analogues. Comp. and Biol. Chem. 2018;76:1–16. doi: 10.1016/j.compbiolchem.2018.05.021. [DOI] [PubMed] [Google Scholar]
- Tian Y., Du D., Rai D., Wang L., Liu H., Zhan P., De Clercq E., Pannecouque C., Liu X. Used heterocyclic compounds bearing bridgehead nitrogen as potent HIV-1 NNRTIs. Part 1: Design, synthesis and biological evaluation of novel 5,7-disubstituted pyrazolo[1,5-a]pyrimidine derivatives. Bioorg. Med. Chem. 2014;22(7):2052–2059. doi: 10.1016/j.bmc.2014.02.029. [DOI] [PubMed] [Google Scholar]
- Tronchet J.M., Seman M. Non nucleoside inhibitors of HIV-1 reverse transcriptase: from the biology of reverse transcriptase to molecular design. Curr. Top. Med. Chem. 2003;3(13):1496–1511. doi: 10.2174/1568026033451754. [DOI] [PubMed] [Google Scholar]
- Van Aalten D.M.F., Findlay J.B.C., Amadei A., Berendsen H.J.C. Essential dynamics of the cellular retinol-binding protein evidence for ligand-induced conformational changes. Protein Eng. Des. Sel. 1995;8:1129–1135. doi: 10.1093/protein/8.11.1129. [DOI] [PubMed] [Google Scholar]
- Venkatachalam C.M., Jiang X., Oldfield T., Waldman M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model. 2003;21:289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- Wang R., Lu Y., Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- Wang C., Greene D., Xiao L., Qi R., Luo R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2018;4(87):1–18. doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav G., Ganguly S. Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities; A mini-review. Eur. J. Med. Chem. 2015;97:419–443. doi: 10.1016/j.ejmech.2014.11.053. [DOI] [PubMed] [Google Scholar]
- Yadav M., Srivastava R., Naaz F., Singh A., Verma R., Singh R.K. In silico studies on new oxathiadiazoles as potential anti-HIV agents. Gen. Rep. 2019;14:87–93. doi: 10.1016/j.genrep.2018.12.004. [DOI] [Google Scholar]
- Zhan P., Liu X. Novel HIV-1 non-nucleoside reverse transcriptase inhibitors: a patent review (2005-2010) Expert Opin. Ther. Pat. 2011;21(5):717–796. doi: 10.1517/13543776.2011.568481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.