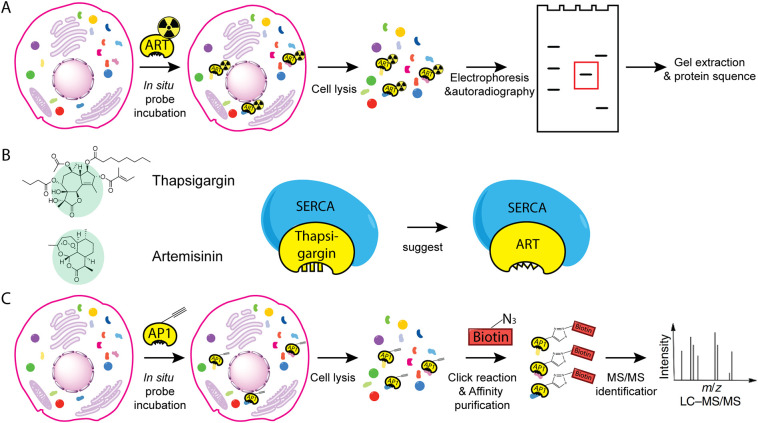
Fig. 2.
General workflow of some methods used to identify targets of artemisinin. (A) Radiolabeling and gel-based protein target identification. The radiolabeled artemisinin was synthesized and used to treat cells. The lysed parasite extracts were separated by electrophoresis. By autoradiography, protein targets of artemisinin were identified and subjected to protein sequencing. (B) Using structural similarity to identify artemisinin targets. Artemisinin and thapsigargin share a similar sesquiterpene lactone structure, thus the target of thapsigargin, SERCA, was proposed to be a target of artemisinin. (C) Using chemical proteomics to identify artemisinin targets. AP1, artemisinin derivative with a clickable alkyne moiety, was used to treat cells for in situ labeling. The lysed parasite extracts were labeled with a biotin moiety through click chemistry before affinity purification and MS-based protein identification.