Fig. 2. OligoFISSEQ-LIT on 36plex-5K.
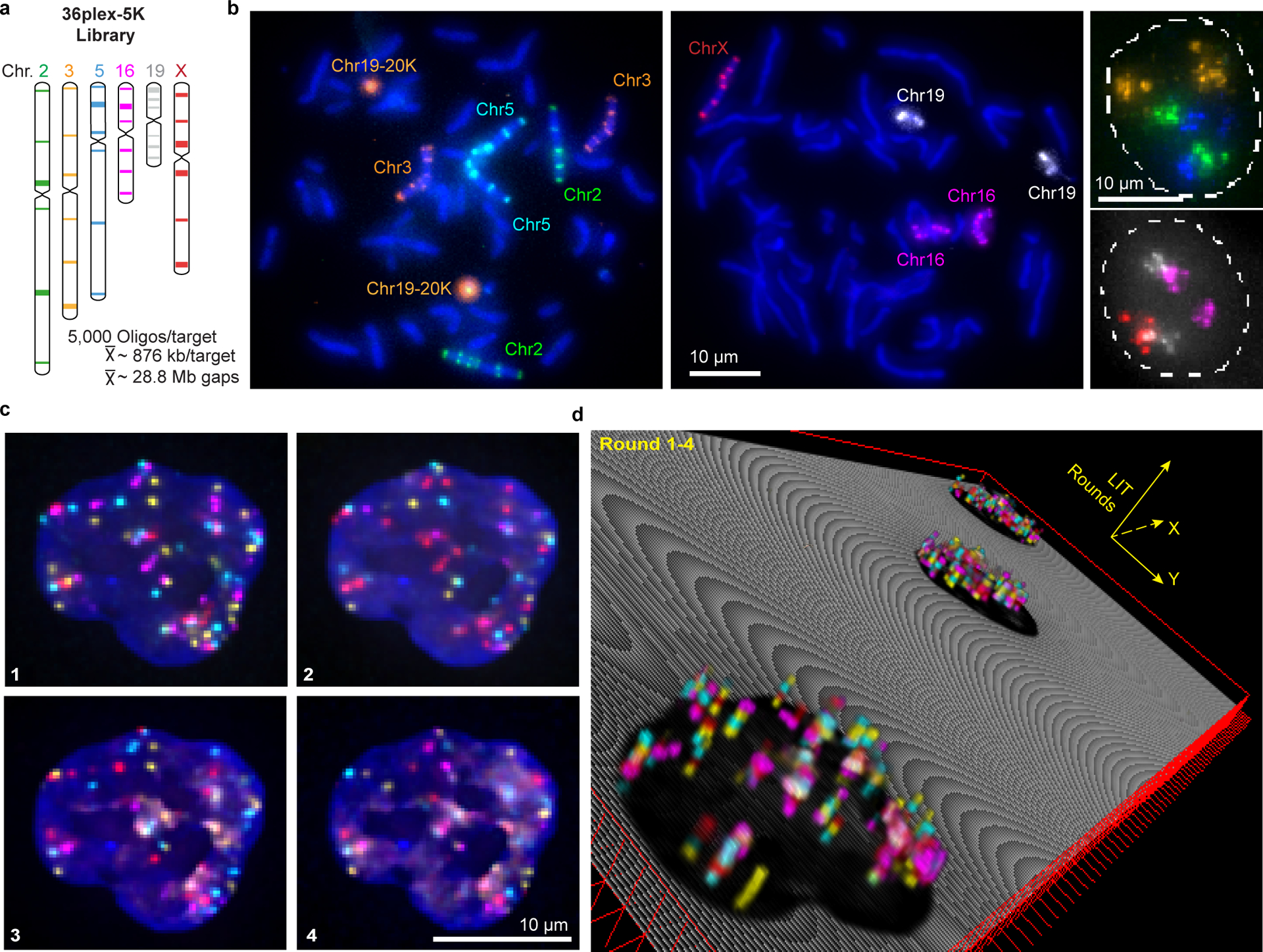
a) Targets of the library. Chromosome number color-coded to correspond to images in panel b. Each target corresponds to a unique barcode.
b) Metaphase chromosome spreads of male lymphoblast cells (left; cells from Applied Genetics; Methods) and interphase nuclei from PGP1f cells (right) representative of four replicates. All six targets on any single chromosome were labeled with secondary oligos carrying the same species (color) of fluorophore. Chr19–20K was used as a positive control in metaphase chromosome spreads. Maximum z-projections.
c) Four rounds of O-LIT off of both streets of 36plex-5K. Images are from deconvolved, five-color merged maximum z-projections. n = 1.
d) 3D representation of a field of view (FOV) containing three cells sequenced with four rounds of O-LIT. Sequencing rounds are represented on the z-axis, with the first being closest to the DAPI-determined nuclear outline (black). Maximum z-projection of sequencing signal from each round was taken, duplicated (2-images total for better visualization), and then stacked on top of each other. The lower left cell corresponds to the cell in panel c.
