Fig. 6. OligoFISSEQ extensions and applications.
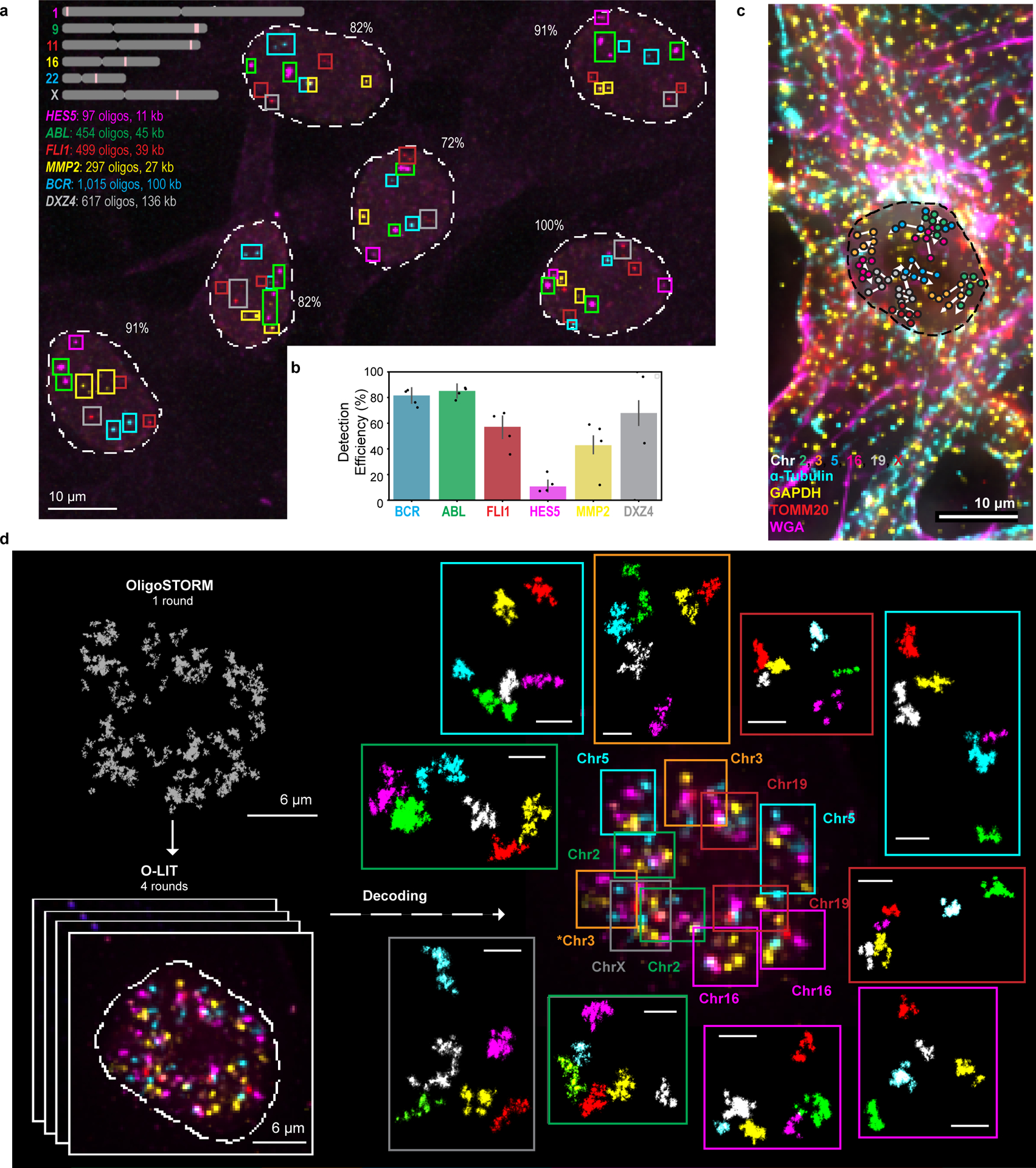
a) O-eLIT detection of single gene targets after sequencing off of both streets. Colored squares mark gene targets identified after 5 rounds of sequencing. Number reflects percentage of targets detected out of 11 (5 autosomal genes x 2 in addition to DXZ4 on the X). Image from deconvolved maximum intensity z-projection and representative of 2 replicates.
b) Tier 1 target detection efficiency from experiment in panel a (n = 61 from 2 replicates). Tier 2 inapplicable due to lack of targets from the same chromosome. Detection efficiency from individual replicates are plotted. Error bars represent 95% bootstrap confidence interval of the mean.
c) Combining O-LIT and immunofluorescence. 36plex-5K was sequenced for four rounds with O-LIT off of both streets, followed by immunofluorescence and staining with wheat germ agglutinin (WGA). Image from deconvolved maximum intensity z projection with chromosome traces overlaid. n = 1.
d) 36plex-5K was hybridized to PGP1f cells and imaged with 1 round of OligoSTORM (2 hours) to visualize all 66 targets simultaneously, followed by 4 rounds of O-LIT (2 – 3 hours per round) to decode targets. Top left, OligoSTORM image showing entire field of view with all unidentified targets. Bottom left, micrograph from first round of O-LIT; image from deconvolved maximum z-projection. Right, all 6 chromosomes identified (central image decorated with colored squares, color coded by chromosome as shown) and then arrayed, in super-resolution, around the central nucleus. All 66 targets excepting one region on Chr16, were detected and identified by O-LIT, with one homolog of Chr3 (*) not captured by OligoSTORM because it happened to fall outside the field of view. All scale bars for OligoSTORM images, 1 μm.
e) Each chromosomal region imaged with OligoSTORM from panel d is displayed separately; orientation may differ from that in panel e. n = 1.
