Abstract
Beneficial microbes are all around us and it remains to be seen, whether all diseases and disorders can be prevented or treated with beneficial microbes. In this study, the presence of various beneficial bacteria were identified from the sediments of Indian major Rivers Ganga and Yamuna from nine different sites using a metagenomic approach. The metagenome sequence analysis using the Kaiju Web server revealed the presence of 69 beneficial bacteria. Phylogenetic analysis among these bacterial species revealed that they were highly diverse. Relative abundance analysis of these bacterial species is highly correlated with different pollution levels among the sampling sites. The PCA analysis revealed that Lactobacillus spp. group of beneficial bacteria are more associated with sediment sampling sites, KAN-2 and ND-3; whereas Bacillus spp. are more associated with sites, FAR-2 and ND-2. This is the first report revealing the richness of beneficial bacteria in the Indian rivers, Ganga and Yamuna. The study might be useful in isolating different important beneficial microorganisms from these river sediments, for possible industrial applications.
Introduction
Rivers are known to be important for the development of human civilization, culture, and welfare. They are one of the crucial components of freshwater ecosystems, maintaining large biodiversity which is vital for sustenance of the terrestrial biome. Since rivers are significant reservoirs of the microbiome, they are relentlessly being explored for the search of de novo microbiota. These bacteria are of greater importance due to their different benefits to humans as well as all other strata of organisms present in the trophic pyramid [1]. It provides its rewarding effects generally through four main mechanisms i.e. enhancement of barrier function, intervention with host pathogens, immunomodulation, and assembly of neurotransmitters [2]. These organisms are gaining increasing importance as functional foods as well as prophylactic, therapeutic, and growth supplements for humans [3–5]. Some of the most common human gut probiotics viz. Lactobacillus and Enterococcus are reported to counteract diabetes, obesity, autoimmune disorder, and cancer through the production of metabolites like short-chain fatty acids [6]. Not only for humans, nowadays, the important microbiome is also being used in agriculture, including veterinary and fisheries, to benefit the animal physiology by improving their internal and external environment [5, 7, 8]. However, in fisheries, the scope of microbial treatment is enormous and the use of the same is gaining day by day. The latest study on Labeo rohita established that dietary administration of a probiotic bacterium, Bacillus aerophilus KADR3, improves the disease resistance and enhances the immunity against Aeromonas hydrophila infection [9]. Similarly, the dietary application of B. amyloliquefaciens CCF7, in L. rohita, challenged with a fish pathogenic bacteria, A. hydrophila MTCC 1739, showed beneficial effects [10]. Though many reports are present on discovering microbiome from natural streams of other countries, there is very insufficient literature available on the same in context to the Indian subcontinent especially in the large riverine ecosystems like Ganga and Yamuna. Therefore, in the present study, the abundance of different beneficial microbiota in the selected stretches of the river Ganga and Yamuna have been identified through the metagenomics approach. The metagenomics study has overcome the problem of culture-oriented microbiological studies associated with different environmental samples and came out as a potential search tool for detailed screening of supportive microbiome species present in an ecosystem [11]. Since the total DNA extracted from an environmental sample is a snapshot of the entire microbial community, metagenomics analysis makes it easier for a comprehensive evaluation of the native microbial ecology [12]. The recent computational advancement and evolution of next-generation sequencing, which can generate millions of sequences at improved cost and speed, make it possible to detect microbial biodiversity easily and their abundance directly from the environmental samples [13–14]. As per our knowledge, this is the first report, presenting an analysis of a large sediment metagenome dataset from these rivers in search of beneficial bacteria.
Materials and methods
Sample collection
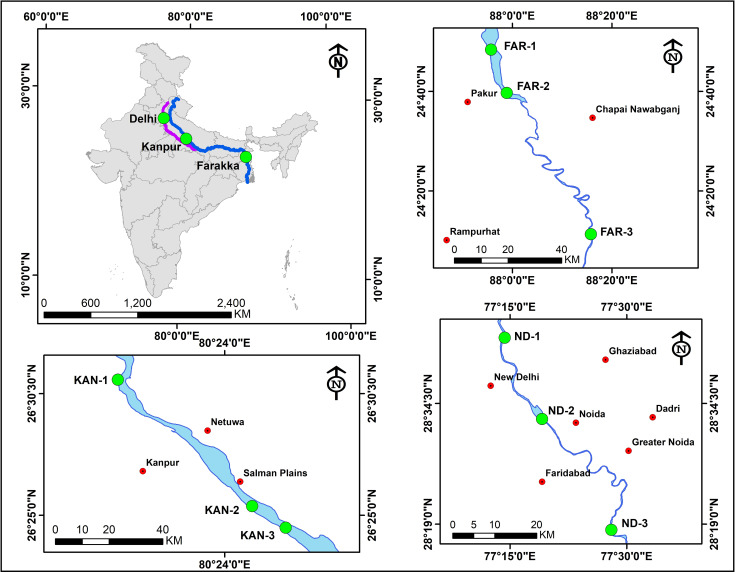
A total of nine sediment samples were collected from the river Ganga and Yamuna. From the Ganga river, six sediment samples were collected from different sites viz. Ganga Barrage (N 26030.858//E 80019.114//) (KAN-1), Jajmau (N 26025.301//E 80025.282//) (KAN-2), Jana Village (N 26024.495//E 80026.904//) (KAN-3) near Kanpur, Uttar Pradesh, Farakka Barrage (N 24047.804//E 87055.417//) (FAR-1), Dhulian (N 24047.804//E 87055.417//) (FAR-2), Lalbagh (N 29011.087//E 88016.079//) (FAR-3) near Farakka, West Bengal. From the river Yamuna, sediment samples were collected from three different sites viz. Wazaribad (N 28042.39//E 77013.57//) (ND-1), Okhla barrage (N 28032.51//E 77018.30//) (ND-2), Faizupur Khaddar (N 28018.43//E 77027.52//) (ND-3) near New Delhi, India (Fig 1).
Fig 1. Map showing the sediment sampling sites.
Sediments were collected from the river Ganga at six locations namely, Kanpur (KAN-1, KAN-2 and KAN-3) and Farakka, (FAR-1, FAR-2 and FAR-3) whereas, three locations from the river Yamuna, at New Delhi (ND-1, ND-2 and ND-3). The map of sediment collection sites was prepared using ArcGIS 10.2.1 platform.
DNA extraction
The obtained samples from different locations from river Ganga and Yamuna were kept in sterile plastic bags, sealed and transported on ice (4°C), and afterward stored at -80°C until further processing. Metagenomic DNA from these sediment samples were extracted using a soil gDNA isolation kit (Nucleospin Soil). After the isolation of metagenomic DNA, the quality was checked in Nanodrop 2000 and Qubit® 3.0 Fluorometer. The metagenomic library was prepared using sufficient amounts of extracted good quality DNA.
Metagenomic library preparation
The paired-end sequencing libraries were arranged using Illumina Trueseq Nano DNA Library Prep Kit. Approximately 200ng of eDNA was fragmented by Covaris M220 to produce a mean fragment allocation of 350bp. Covaris shearing produced dsDNA fragments with 3' or 5' overhangs. The fragments were then subjected to end-repair. As per the description in the kit, the products were PCR amplified with the index primer. The D1000 Screen tape was used to investigate the PCR enriched libraries in the 4200 Tape Station system (Agilent Technologies).
Whole metagenome sequencing and quality assessment
After obtaining the mean peak size from Agilent Tape Station profile and Qubit concentration for the libraries, PE Illumina libraries were loaded into NextSeq 500 for cluster generation and sequencing. After trimming, a minimum length of 100 nt was applied. The CLC Genomics Workbench version 8.5.1 (CLC bio; https://www.qiagenbioinformatics.com/products/clc-genomics-workbench) was used to assemble the filtered high-quality reads of each sample into scaffolds.
Metagenomic sequences accession numbers
The Metagenomic sequences used in this study have been submitted to the NCBI-SRA database under Accession Nos: SRP190174, SRP190175, SRP189880, SRP191076, SRP191079, SRP191075, SRP191073, SRP191080, and SRP191499 for three Kanpur samples (KAN-1, KAN-2, KAN-3), three Farakka samples (FAR-1, FAR-2, FAR-3) and three New Delhi samples (ND-1, ND-2, ND-3) respectively.
Sequence annotation and bioinformatics analysis
For the detection of the beneficial microbiome in the sediment metagenome, filtered metagenomic reads were used for taxonomical binning by the Kaiju web interface. Classifier-Kaiju used Burrows-Wheeler transform algorithm for taxonomic classification on the protein-level [15]. On the other hand, to highlight the phylogenetic relationship among the beneficial microbiome species found in the sediment metagenome, multiple sequence analysis was carried out using MEGA 6 software. The Neighbor-Joining method was used to infer evolutionary history [16]. The Maximum Composite Likelihood method [17] was used to compute evolutionary distances. To understand the evolutionary relationship among the 69 identified beneficial microbial species, derived from the sediments of the rivers, Ganga and Yamuna, a multiple sequence analysis (MSA) was carried out using MEGA 6 software [18]. Relative abundance of beneficial bacteria was calculated using Kaiju Web Server. Comparison was done based on standard student t-test [19]. Heat map presentation was arranged using multiple experiment viewer (MeV), a standalone tool for visualizing the clustering of multivariate data [20]. The Principal Component Analysis (PCA) biplot and Scatterplot matrix along with correlation values between sampling sites and relative abundance of helpful bacteria were developed in JMP Pro 10 after the standardization of the estimated data.
Results
Sequence generation
Sediment samples from nine sites (Fig 1) of river Ganga (KAN-1, KAN-2, and KAN-3; FAR-1, FAR-2, and FAR-3) and river Yamuna (ND-1, ND-2, and ND-3) were analyzed using high throughput next-generation sequencing to identify the microbial biodiversity. The total number of high quality reads with their consequent data volume of each sediment samples are presented in Table 1. All the high quality reads obtained from the sediments of different sites were considered for sensitive taxonomic classification analysis. However, the taxonomic classification study could not classify all the reads. Only 41.58%, 49.35%, and 54.79% of the total reads were classified form Kanpur sediment samples (KAN-1, KAN-2, and KAN-3) of river Ganga, respectively. In Farakka sediment samples (FAR-1, FAR-2, and FAR-3) of river Ganga, only 50.82%, 52.08%, and 35.68% of the total reads were classified. Similarly, in New Delhi sediment samples (ND-1, ND-2, and ND-3) of river Yamuna, 53.37%, 38.95%, and 44.82% of the total reads were classified, respectively.
Table 1. Data details of nine sediment metagenome of river Ganga and Yamuna.
| Location | Description | High quality reads (bp) |
|---|---|---|
| KAN-1 | 28,718,955 | |
| Kanpur | KAN-2 | 33,703,138 |
| KAN-3 | 33,887,572 | |
| FAR-1 | 24,929,338 | |
| Farakka | FAR-2 | 29,128,182 |
| FAR-3 | 54,496,302 | |
| ND-1 | 64,876,611 | |
| New Delhi | ND-2 | 64,749,798 |
| ND-3 | 62,670,420 |
Taxonomical classification of sediment metagenome
Based on the taxonomical classification, a large number of beneficial bacterial species (Table 2) were identified from the sediment samples of the rivers, Ganga, and Yamuna. Four Vibrio (V. mediterranei, V. fluvialis, V. gazogenes, and V. alginolyticus), nine Bacillus (B. clausii, B. circulans, B. subtilis, B. coagulans, B. cereus, B. megaterium, B. mycoides, B. pumilus, and B. licheniformis), sixteen Lactobacillus (L. curvatus, L. brevis, L. casei, L. acidophilus, L. buchneri, L. crispatus, L. delbrueckii, L. fermentum, L. gasseri, L. helveticus, L. johnsonii, L. paracasei, L. plantarum, L. reuteri, L. rhamnosus and L. salivarius), five Bifidobacterium (B. animalis, B. bifidum, B. longum, B. breve, and B. adolescentis), three Shewanella (S. colwelliana, S. putrefaciens and S. xiamenensis), three Pediococcus (P. pentosaceus, P. Acidilactici and P. ethanolidurans), six Enterococcus (E. durans, E. faecium, E. faecalis E. raffinosus E. hirae and E. mundtii), four Pseudomonas (P. fluorescens, P. chlororaphis, P. stutzeri and P. synxantha), four Roseobacter (R. litoralis, R. denitrificans, R. litoralis and R. denitrificans), four Oenococcus (O. oeni, O. kitaharae, O. alcoholitolerans and O. oeni AWRIB429), two Carnobacterium (C. divergens and C. maltaromaticum), two Streptococcus (S. salivarius and S. thermophilus), two Vagococcus (V. fluvialis bH819 and V. teuberi) along with one each for, Aeromonas veronii, Leuconostocme senteroides, Micrococcus luteus, Paenibacillus polymyxa, and Lactococcus lactis were identified from the metagenome.
Table 2. Relative abundance of beneficial bacteria species identified from the nine sediment metagenome of river Ganga and Yamuna.
| Name of the Genus | Name of the species | KAN-1 | KAN-2 | KAN-3 | Average | SD | FAR-1 | FAR-2 | FAR-3 | Average | SD | ND-1 | ND-2 | ND-3 | Average | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. curvatus | 0.00008 | 0.0002 | 0.00002 | 1.0E-04 | 9.17E-05 | 0.0001 | 0.0001 | 0.00009 | 9.7E-05 | 5.77E-06 | 0.0001 | 0.0001 | 0.0002 | 1.3E-04 | 5.77E-05 | |
| L. brevis | 0.0001 | 0.0003 | 0.0002 | 2.0E-04 | 1.00E-04 | 0.0005 | 0.0003 | 0.0006 | 4.7E-04 | 1.5E-04 | 0.0003 | 0.0003 | 0.0003 | 3.0E-04 | 0 | |
| L. casei | 0.0004 | 0.0006 | 0.0002 | 4E-04 | 2.00E-04 | 0.0008 | 0.0005 | 0.0004 | 5.7E-04* | 2.1E-04 | 0.0001 | 0.0001 | 0.0001 | 1.0E-04* | 0 | |
| L. acidophilus | 0.00003 | 0.00004 | 0.00002 | 3E-05 | 1.00E-05 | 0.00009 | 0.00003 | 0.00004 | 5.33E-05 | 3.21E-05 | 0.00005 | 0.00004 | 0.00007 | 5.33E-05 | 1.52E-05 | |
| L. buchneri | 0.00004 | 0.00009 | 0.00003 | 5.3E-05 | 3.2E-05 | 0.00009 | 0.00004 | 0.00008 | 7.0E-05 | 2.65E-05 | 0.00006 | 0.00008 | 0.00009 | 7.67E-05 | 1.52E-05 | |
| L. crispatus | 0.0002 | 0.0004 | 0.0002 | 2.7E-04 | 1.2E-04 | 0.0003 | 0.0003 | 0.0002 | 2.7E-04 | 5.7E-05 | 0.0003 | 0.0002 | 0.0004 | 3.0E-04 | 1.0E-04 | |
| L. delbrueckii | 0.0001 | 0.0003 | 0.0001 | 1.7E-04 | 1.2E-04 | 0.0003 | 0.0001 | 0.0001 | 1.7E-04 | 1.1E-04 | 0.0003 | 0.0003 | 0.0006 | 04.0E-04 | 1.7E-04 | |
| L. fermentum | 0.0002 | 0.0003 | 0.00007 | 1.9E-04 | 1.2E-04 | 0.0003 | 0.0002 | 0.0002 | 2.3E-04 | 5.77E-05 | 0.0002 | 0.0003 | 0.0004 | 3.0E-04 | 1.0E-04 | |
| L. gasseri | 0.0001 | 0.0003 | 0.0001 | 1.7E-04 | 1.2E-04 | 0.0002 | 0.0001 | 0.00006 | 1.2E-04 | 7.21E-05 | 0.0002 | 0.0001 | 0.0003 | 2.0E-04 | 1.0E-04 | |
| L. helveticus | 0.00005 | 0.0001 | 0.00005 | 6.67E-05 | 2.88E-05 | 0.0001 | 0.00008 | 0.00006 | 8.0E-05 | 2.0E-05 | 0.0001 | 0.00008 | 0.0002 | 1.3E-04 | 6.4E-05 | |
| Lactobacillus | L. johnsonii | 0.00007 | 0.0001 | 0.00003 | 6.67E-05 | 3.5E-05 | 0.0001 | 0.0001 | 0.00007 | 9.0E-05 | 1.73E-05 | 0.0001 | 0.0001 | 0.0001 | 1.0E-04 | 0 |
| L. paracasei | 0.0001 | 0.0002 | 0.00008 | 1.3E-04 | 6.43E-05 | 0.0002 | 0.0001 | 0.0001 | 1.3E-04 | 5.77E-05 | 0.0001 | 0.0001 | 0.0002 | 1.3E-04 | 5.7E-05 | |
| L. plantarum | 0.0003 | 0.0005 | 0.0003 | 3.7E-04 | 1.2E-04 | 0.0004 | 0.0003 | 0.0003 | 3.3E-04 | 5.77E-05 | 0.0006 | 0.0004 | 0.0007 | 5.7E-04 | 1.5E-04 | |
| L. reuteri | 0.0003 | 0.0006 | 0.0002 | 3.7E-04 | 2.1E-04 | 0.0004 | 0.0003 | 0.0002 | 3.0E-04 | 1.0E-04 | 0.0006 | 0.0004 | 0.0007 | 5.7E-04 | 1.5E-04 | |
| L. rhamnosus | 0.0002 | 0.0004 | 0.0001 | 2.3E-04 | 1.5E-04 | 0.0004 | 0.0002 | 0.0001 | 2.3E-04 | 1.5E-04 | 0.0002 | 0.0002 | 0.0003 | 2.3E-04 | 5.7E-05 | |
| L. salivarius | 0.0003 | 0.0006 | 0.0002 | 3.7E-04 | 2.1E-04 | 0.0005 | 0.0003 | 0.0002 | 3.3E-04 | 1.5E-04 | 0.001 | 0.0005 | 0.001 | 8.3E-04 | 2.9E-04 | |
| B. clausii | 0.001 | 0.001 | 0.0005 | 8.3E-04 | 2.89E-04 | 0.002 | 0.001 | 0.001 | 1.3E-03* | 5.7E-04 | 0.0002 | 0.0004 | 0.0004 | 3.3E-04* | 1.2E-04 | |
| B. circulans | 0.0003 | 0.0006 | 0.0002 | 3.7E-04 | 2.08E-04 | 0.0005 | 0.0004 | 0.0003 | 4.0E-04 | 1.0E-04 | 0.0002 | 0.0004 | 0.0003 | 3.0E-04 | 1.0E-04 | |
| Bacillus | B. subtilis | 0.0006 | 0.0008 | 0.0004 | 6.0E-04 | 2.00E-04 | 0.001 | 0.0007 | 0.0006 | 7.7E-04 | 2.0E-04 | 0.0003 | 0.0008 | 0.0007 | 6.0E-04 | 2.6E-04 |
| B. coagulans | 0.001 | 0.002 | 0.0006 | 1.2E-03 | 7.21E-04 | 0.002 | 0.001 | 0.001 | 1.3E-03 | 5.7E-04 | 0.001 | 0.001 | 0.001 | 1.0E-03 | 0 | |
| B. cereus | 0.003 | 0.004 | 0.002 | 3.0E-03 | 1.00E-03 | 0.006 | 0.003 | 0.007 | 5.3E-03 | 2.1E-03 | 0.003 | 0.004 | 0.004 | 3.7E-03 | 5.8E-04 | |
| B. megaterium | 0.0006 | 0.001 | 0.0003 | 6.3E-04 | 3.51E-04 | 0.0009 | 0.0009 | 0.001 | 9.3E-04 | 5.77E-05 | 0.0005 | 0.001 | 0.0008 | 7.7E-04 | 2.5E-04 | |
| B. mycoides | 0.0005 | 0.0006 | 0.0007 | 6.0E-04* | 1.00E-04 | 0.0006 | 0.0005 | 0.0004 | 5.0E-04 | 1.0E-04 | 0.0002 | 0.0004 | 0.0004 | 3.3E-04* | 1.2E-04 | |
| B. pumilus | 0.0004 | 0.0006 | 0.0002 | 4.0E-04 | 2.00E-04 | 0.0008 | 0.0006 | 0.0004 | 6.0E-04 | 2.0E-04 | 0.0003 | 0.0008 | 0.0007 | 6.0E-04 | 2.6E-04 | |
| B. licheniformis | 0.0002 | 0.0002 | 0.0001 | 1.07E-04 | 5.77E-05 | 0.0003 | 0.0002 | 0.0001 | 2.0E-04 | 1.0E-04 | 0.0001 | 0.0002 | 0.0002 | 1.7E-04 | 5.77E-05 | |
| P. pentosaceus | 0.00007 | 0.0002 | 0.00006 | 1.1E-04 | 7.81E-05 | 0.0002 | 0.0002 | 0.0001 | 1.7E-04 | 5.77E-05 | 0.0003 | 0.0001 | 0.0002 | 2.0E-04 | 1.0E-04 | |
| Pediococcus | P. acidilactici | 0.008 | 0.008 | 0.1 | 3.9E-02 | 5.31E-02 | 0.0003 | 0.0003 | 0.0002 | 2.7E-04 | 5.77E-05 | 0.0003 | 0.0002 | 0.0003 | 2.7E-04 | 5.77E-05 |
| P. ethanolidurans | 0.00007 | 0.00008 | 0.00005 | 6.67E-05 | 1.53E-05 | 0.00007 | 0.00007 | 0.00004 | 6.0E-05 | 1.73E-05 | 0.00006 | 0.00006 | 0.00009 | 7.0E-05 | 1.73E-05 | |
| V. mediterranei | 0.00005 | 0.00004 | 0.00005 | 4.67E-05 | 5.77E-06 | 0.0001 | 0.00004 | 0.00005 | 6.3E-05 | 3.2E-05 | 0.00003 | 0.00005 | 0.00005 | 4.3E-05 | 1.15E-05 | |
| Vibrio | V. fluvialis | 0.0002 | 0.0002 | 0.0003 | 2.3E-04 | 5.77E-05 | 0.0002 | 0.0002 | 0.0002 | 2.0E-04 | 0 | 0.0004 | 0.0001 | 0.0002 | 2.3E-04 | 1.5E-04 |
| V. gazogenes | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | 0.0003 | 0.0002 | 0.0003 | 2.7E-04** | 5.77E-05 | |
| V. alginolyticus | 0.0007 | 0.0008 | 0.0009 | 8.0E-04 | 1.00E-04 | 0.0009 | 0.0008 | 0.0005 | 7.3E-04 | 2.1E-04 | 0.001 | 0.0006 | 0.001 | 8.7E-04 | 2.3E-04 | |
| R. litoralis | 0.001 | 0.001 | 0.003 | 1.6E-03 | 1.18E-03 | 0.0009 | 0.0006 | 0.0007 | 7.3E-04 | 1.5E-04 | 0.0020 | 0.0004 | 0.0005 | 9.7E-04 | 8.9E-04 | |
| Roseobacter | R. denitrificans | 0.001 | 0.0009 | 0.003 | 1.6E-03 | 1.18E-03 | 0.0007 | 0.0006 | 0.0006 | 6.3E-04 | 5.77E-05 | 0.0020 | 0.0004 | 0.0004 | 9.3E-04 | 9.2E-04 |
| R. litoralis 149 | 0.00002 | 0.00003 | 0.00009 | 4.67E-05 | 3.79E-05 | 0.00002 | 0.00001 | 0.00001 | 1.3E-05 | 5.77E-06 | 0.000008 | 0.00001 | 0.000005 | 7.7E-06 | 2.5E-06 | |
| R. denitrificans 114 | 0.00006 | 0.00006 | 0.0002 | 1.1E-04 | 8.08E-05 | 0.00003 | 0.00003 | 0.00002 | 2.7E-05 | 5.77E-06 | 0.00008 | 0.00002 | 0.00003 | 4.3E-05 | 3.2E-05 | |
| Vagococcus | V. fluvialis bH819 | 0 | 0 | 0 | 0* | 0.00E+00 | 0 | 0 | 0.000002 | 6.7E-07* | 1.15E-06 | 0.0003 | 0.0004 | 0.0006 | 4.3E-04** | 1.5E-04 |
| V. teuberi | 0.0002 | 0.0005 | 0.0001 | 2.7E-04 | 2.08E-04 | 0 | 0.0003 | 0.0002 | 1.7E-04 | 1.5E-04 | 0.00006 | 0.00005 | 0.0001 | 7.0E-05 | 2.6E-05 | |
| O. oeni | 0.0003 | 0.0004 | 0.0002 | 3.0E-04 | 1.00E-04 | 0.0004 | 0.0003 | 0.0002 | 3.0E-04 | 1.0E-04 | 0.0003 | 0.0003 | 0.0004 | 3.3E-04 | 5.77E-05 | |
| Oenococcus | O. kitaharae | 0.0002 | 0.0003 | 0.0001 | 2.0E-04 | 1.00E-04 | 0.0004 | 0.0002 | 0.0002 | 2.7E-04 | 1.1E-04 | 0.0001 | 0.0002 | 0.0002 | 1.7E-04 | 5.77E-05 |
| O. alcoholitolerans | 0.0001 | 0.0001 | 0.00006 | 8.67E-05 | 2.31E-05 | 0.0002 | 0.0001 | 0.00009 | 1.3E-04 | 6.1E-05 | 0.00008 | 0.0001 | 0.0002 | 1.3E-04 | 6.4E-05 | |
| O. oeni AWRIB429 | 0.00001 | 0.000006 | 0.000003 | 6.33E-06 | 3.51E-06 | 0.00002 | 0.00001 | 0.000004 | 1.1E-05 | 8.1E-06 | 0 | 0.000003 | 0.000002 | 1.67E-06 | 1.5E-06 | |
| P. fluorescens | 0.04 | 0.01 | 0.02 | 2.3E-02 | 1.53E-02 | 0.01 | 0.008 | 0.008 | 8.7E-03 | 1.2E-03 | 0.01 | 0.005 | 0.007 | 7.3E-03 | 2.5E-03 | |
| Pseudomonas | P. chlororaphis | 0.002 | 0.002 | 0.003 | 2.3E-03 | 5.77E-04 | 0.002 | 0.002 | 0.001 | 1.7E-03 | 5.7E-04 | 0.002 | 0.001 | 0.001 | 1.3E-03 | 5.8E-04 |
| P. stutzeri | 0.008 | 0.02 | 0.02 | 1.6E-02 | 6.93E-03 | 0.008 | 0.006 | 0.006 | 6.7E-03 | 1.2E-03 | 0.02 | 0.005 | 0.009 | 1.13E-02 | 7.8E-03 | |
| P. synxantha | 0.0002 | 0.0002 | 0.0003 | 2.3E-04 | 5.77E-05 | 0.0002 | 0.0001 | 0.0001 | 1.3E-04 | 5.7E-05 | 0.0003 | 0.0001 | 0.0001 | 1.7E-04 | 1.2E-04 | |
| S. colwelliana | 0.0008 | 0.0006 | 0.0009 | 7.7E-04* | 1.53E-04 | 0.0007 | 0.0005 | 0.0005 | 5.7E-04 | 1.2E-04 | 0.0005 | 0.0004 | 0.0005 | 4.7E-04* | 5.77E-05 | |
| Shewanella | S. putrefaciens | 0.0003 | 0.0007 | 0.006 | 2.3E-03 | 3.18E-03 | 0.0003 | 0.0002 | 0.0002 | 2.3E-04 | 5.77E-05 | 0.0004 | 0.0002 | 0.0003 | 3.0E-04 | 1.0E-04 |
| S. xiamenensis | 0.0001 | 0.0003 | 0.0004 | 2.6-E04 | 1.53E-04 | 0.0002 | 0.0001 | 0.0001 | 1.3E-04 | 5.77E-05 | 0.0004 | 0.0001 | 0.0002 | 2.3E-04 | 1.5E-04 | |
| E. durans | 0.00009 | 0.0002 | 0.00005 | 1.1E-04 | 7.77E-05 | 0.0001 | 0.0001 | 0.0001 | 1.0E-04 | 0 | 0.0001 | 0.0002 | 0.0003 | 2.0E-04 | 1.0E-04 | |
| E. faecium | 0.0003 | 0.001 | 0.0002 | 5.0E-04 | 4.36E-04 | 0.0006 | 0.0003 | 0.0004 | 4.3E-04* | 1.5E-04 | 0.001 | 0.0006 | 0.001 | 8.7E-04* | 2.3E-04 | |
| Enterococcus | E. faecalis | 0.0006 | 0.002 | 0.0005 | 1.0E-03 | 8.39E-04 | 0.0008 | 0.0006 | 0.0005 | 6.3E-04 | 1.5E-04 | 0.001 | 0.0009 | 0.002 | 1.3E-03 | 6.1E-04 |
| E. raffinosus | 0.00001 | 0.00006 | 0.00001 | 2.67E-05 | 2.89E-05 | 0.00002 | 0.00001 | 0.00002 | 1.7E-05 | 5.77E-06 | 0.00004 | 0.00002 | 0.00005 | 3.67E-05 | 1.5E-05 | |
| E. hirae | 0.00002 | 0.00009 | 0.00001 | 4.0E-05 | 4.36E-05 | 0.00007 | 0.00003 | 0.00003 | 4.3E-05 | 2.3E-05 | 0.0006 | 0.00007 | 0.0001 | 2.7E-04 | 3.0E-04 | |
| E. mundtii | 0.0001 | 0.0003 | 0.00009 | 1.6E-04 | 1.18E-04 | 0.0002 | 0.0001 | 0.0001 | 1.3E-04 | 5.77E-05 | 0.0002 | 0.0002 | 0.0002 | 2.0E-04 | 0 | |
| B. animalis | 0.0003 | 0.0004 | 0.0002 | 3.0E-04 | 1.00E-04 | 0.0004 | 0.0003 | 0.0002 | 3.0E-04 | 1.0E-04 | 0.0005 | 0.0002 | 0.0003 | 3.3E-04 | 1.5E-04 | |
| Bifidobacterium | B. bifidum | 0.0002 | 0.0004 | 0.0001 | 2.3E-04 | 1.53E-04 | 0.0003 | 0.0002 | 0.0002 | 2.3E-04 | 5.7E-05 | 0.003 | 0.0003 | 0.0008 | 1.4E-03 | 1.4E-03 |
| B. longum | 0.0004 | 0.002 | 0.0004 | 9.3E-04 | 9.24E-04 | 0.0004 | 0.0004 | 0.0004 | 4.0E-04 | 0 | 0.01 | 0.001 | 0.003 | 4.7E-03 | 4.7E-03 | |
| B. breve | 0.0002 | 0.0005 | 0.0002 | 3.0E-04 | 1.73E-04 | 0.0004 | 0.0002 | 0.0002 | 2.7E-04 | 1.2E-04 | 0.002 | 0.0003 | 0.0008 | 1.0E-03 | 8.7E-04 | |
| B. adolescentis | 0.0004 | 0.004 | 0.0007 | 2.0E-03 | 2.0E-03 | 0.0004 | 0.0003 | 0.0003 | 3.0E-04 | 1.0E-04 | 0.03 | 0.002 | 0.009 | 1.4E-02 | 1.5E-02 | |
| Carnobacterium | C. divergens | 0.0003 | 0.0006 | 0.0002 | 3.7E-04 | 2.1E-04 | 0.0003 | 0.0003 | 0.0002 | 2.7E-04 | 5.77E-05 | 0.0002 | 0.0003 | 0.0004 | 3.0E-04 | 1.0E-04 |
| C. maltaromaticum | 0.0004 | 0.001 | 0.0003 | 5.7E-04 | 3.8E-04 | 0.0006 | 0.0004 | 0.0004 | 4.7E-04 | 1.2E-04 | 0.0005 | 0.0004 | 0.0007 | 5.0E-04 | 1.5E-04 | |
| Lactococcus | L. lactis | 0.0007 | 0.002 | 0.0004 | 1.03E-03 | 8.5E-04 | 0.001 | 0.0008 | 0.0006 | 8.0E-04 | 2.0E-04 | 0.001 | 0.0009 | 0.002 | 1.3E-03 | 6.0E-04 |
| Leuconostock | L. mesenteroides | 0.0002 | 0.0005 | 0.002 | 9.0E-04 | 9.6E-04 | 0.0003 | 0.0002 | 0.0002 | 2.3E-04 | 5.77E-05 | 0.0007 | 0.0003 | 0.001 | 6.7E-04 | 3.5E-04 |
| Micrococcus | M. luteus | 0.0007 | 0.003 | 0.0009 | 1.5E-03 | 1.3E-03 | 0.002 | 0.001 | 0.0008 | 1.3E-03 | 6.4E-04 | 0.001 | 0.0004 | 0.0006 | 6.7E-04 | 3.0E-04 |
| Streptococcus | S. salivarius | 0.00007 | 0.0003 | 0.00002 | 1.3E-04 | 1.5E-04 | 0.00009 | 0.00004 | 0.00005 | 6.0E-05 | 2.64E-05 | 0.0004 | 0.0001 | 0.0004 | 3.0E-04 | 1.7E-04 |
| S. thermophilus | 0.00009 | 0.0003 | 0.00005 | 1.5E-04 | 1.3E-04 | 0.0001 | 0.00008 | 0.00007 | 8.3E-05** | 1.5E-05 | 0.0003 | 0.0003 | 0.0004 | 3.0E-04** | 5.77E-05 | |
| Paenibacillus | P. polymyxa | 0.0009 | 0.001 | 0.0005 | 8E-04 | 2.6E-04 | 0.001 | 0.001 | 0.0008 | 9.0E-04 | 1.2E-04 | 0.0008 | 0.001 | 0.001 | 9.3E-04 | 1.2E-04 |
| Aeromonas | A. veronii | 0.001 | 0.002 | 0.004 | 2.3E-03 | 1.5E-03 | 0.0008 | 0.0006 | 0.0006 | 6.7E-04 | 1.2E-04 | 0.002 | 0.0006 | 0.0009 | 1.2E-03 | 7.3E-04 |
p≤0.05 * p≤0.01** denote the level significance in Student t-test among average value in the respective row.
Phylogenetic analysis
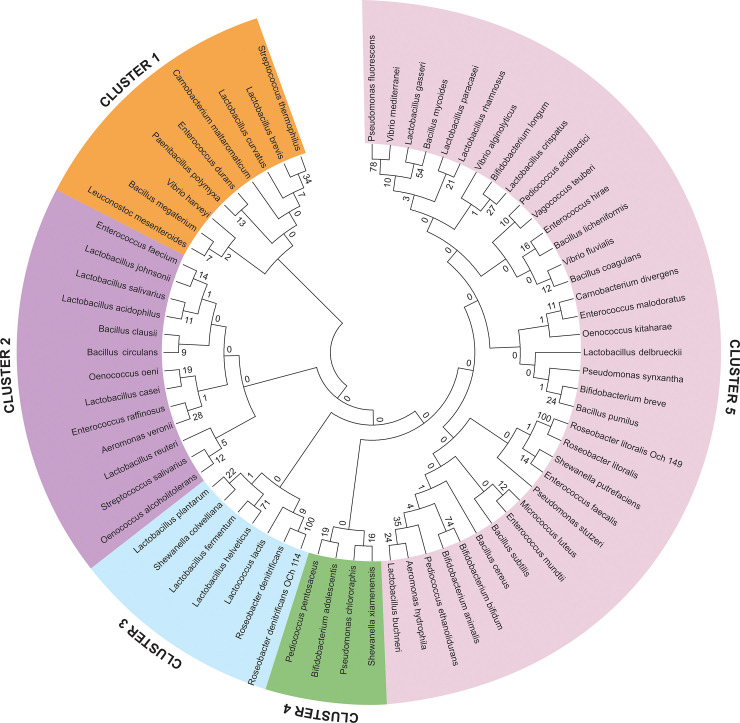
MSA revealed that the majority of the species showed diversity. Phylogenetic tree analysis delineated that, all the species shaped five different clusters (Fig 2). In the first CLUSTER, S. thermophilus and L. brevis derived from Yamuna and Farakka sediment samples respectively were found phylogenetically very close to each other with the bootstrap value of 34. In CLUSTER-2, E. faecium and L. johnsonii, derived from Yamuna and Farakka sediment samples respectively, were found very close to each other with a bootstrap value of 14. Similarly, in CLUSTER-3, L. fermentum and L. helveticus derived from Kanpur and Yamuna sediment samples respectively, were found phylogenetically related with a high bootstrap value of 71. In CLUSTER-4, P. pentosaceus and B. adolescentis both derived from Yamuna sediment samples were found close to each other with the bootstrap value of 19. The highest numbers of evolutionary closed beneficial microbiome species were found in CLUSTER-5. L. gasser and B. mycoides derived from Kanpur and Yamuna sediment samples were found close to each other with a bootstrap value of 54.
Fig 2. Phylogenetic tree of 69 identified genome of helpful bacterial species derived from the sediments metagenome of the river Ganga and Yamuna.
Relative abundance at different sites
In the classified metagenomics data, a total of 69 species of the bacteria from 18 different genera were considered for analysis. Heat map analysis showed a clear distinction in the relative abundance of different bacteria between Kanpur and Farakka sediment samples of river Ganga. Similarly, the prevalence of beneficial bacterial species in the sediment samples of river Yamuna was also different from Kanpur and Farakka stretches of river Ganga (Fig 3).
Fig 3. Heat map of relative species abundance of identified beneficial bacteria from nine different sediment metagenome.
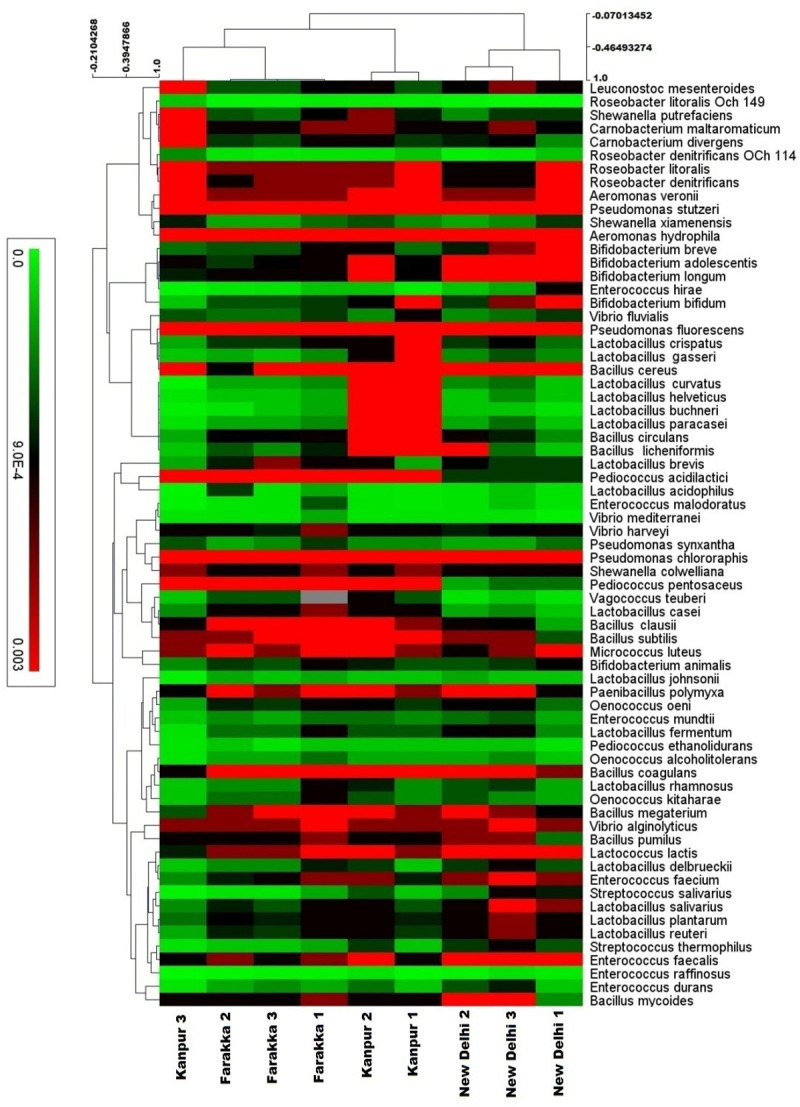
Heat map showing 69 species of beneficial bacteria with significant differences of relative abundances among the nine sampling sites of river Ganga and Yamuna.
Relative abundance analysis revealed that the species L. curvetus and L. brevis were present in similar proportion in sediment samples of all the nine sampling sites of the two rivers; however, L. casei was present in relatively high proportion at Farakka stretch of river Ganga with statistical significance (p-value of 0.02). B. clausii was found in a high proportion (p≤0.05) at Farakka stretch whereas, B. mycoides found in a high proportion (p≤0.05) at Kanpur stretch of river Ganga. Our metagenomic data showed that, one species of Vibrio (V. harveyi) which showed differential relative abundance between three locations (Kanpur, Farakka and New Delhi) and was found relatively lower (p≤0.05) proportion at New Delhi stretch of river Yamuna as compared to Kanpur stretch of river Ganga. Similarly, S. colwelliana was found in a higher proportion (p≤0.05) at Kanpur stretch of river Ganga. E. faecium was found in high proportion at New Delhi stretch of river Yamuna as compared to other locations (p≤0.05) (Table 2).
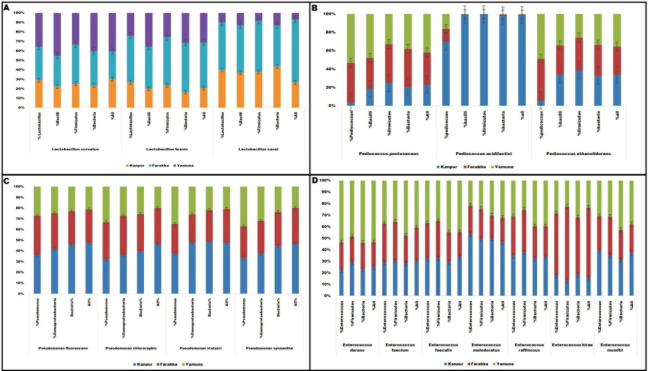
Based on the taxonomical hierarchy, it was revealed that, in all the three locations (Kanpur, Farakka, and New Delhi), L. curvatus had similar relative abundances. The species, L. brevis also showed a similar trend, however, its relative abundance was comparatively higher in the sediment samples of Farakka stretch of river Ganga. The L. casei showed lower abundance in sediment samples at New Delhi stretch of river Yamuna as compared to the other two sites of river Ganga (Fig 4A). Among the Pediococcus population, it is interesting to note that, in the sediment metagenome of Kanpur site of river Ganga, the P. acidilactici was (Student’s t-test, p ≤0.05) dominant over the all taxonomical profile; however, P. pentosaceus and P. ethanolidurans showed equal distribution among the sediment metagenomes at Farakka of river Ganga and New Delhi of river Yamuna (Fig 4B). Likewise, Pseudomonas population showed an equal distribution of relative abundance in all the nine sites. However, P. fluorescens, P. chlororaphis showed (Student’s t-test, p ≤0.05) relative abundance value at Kanpur (Fig 4C). Among the Enterococcus spp., E. durans, E. malodoratus, E. raffinosus, E. hirae, and E. mundtii showed non-significant differences among the nine sampling sites. E. faecium and E. faecalis showed higher abundance (Student’s t-test, p ≤0.05) in sediment metagenomes of river Yamuna compared to Kanpur and Farakka stretch of river Ganga (Fig 4D).
Fig 4. Relative abundance of beneficial microbes on the basis of their taxonomical profile, where A, B, C, and D represent Lactobacillus, Prediococcus, Pseudomonas and Enterococcus groups of different beneficial microbes respectively.
(A) L. casei showed significantly (Student’s t-test, p ≤0.05) lower abundance in New Delhi as compared to rest two locations. (B) Prediococcus pentosaceus and Prediococcus ethanolidurans showed equal abundance among the Farakka and New Delhi locations. Prediococcus acidilacticiis significantly dominant over the all taxonomical profile at Kanpur (Student’s t-test, p ≤0.05). (C) Found significant difference (Student’s t-test, p ≤0.05) for Pseudomonas fluorescens, Pseudomonas chlororaphis. (D) Enterococcus faecium and Enterococcus faecalis showed higher abundance (Student’s t-test, p ≤0.05) in Kanpur and Farakka, respectively.
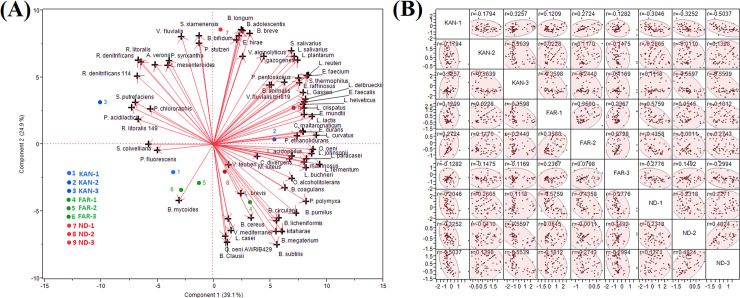
The biplot of principal component analysis (PCA), the PC1, and PC2 altogether could explain 64% variability in the data which showed that the sites at Farraka are closely associated and sites at Kanpur and New Delhi are diverse about the relative abundance of beneficial bacteria (Fig 5A). The relative abundance of beneficial bacteria is found to be closely associated at site FAR-1, KAN-2, KAN-3, ND-1, ND-2, and ND-3. Further, PCA showed that Lactobacillus spp. group of beneficial bacteria are more associated with sites KAN-2 and ND-3; whereas Bacillus spp. are more associated with FAR-2 and ND-2. The Scatter plot matrix showed the correlation between the sites about the relative abundance of beneficial bacteria (Fig 5B). Highest positive correlation was found between ND-2 and ND-3 (r = 0.48) followed by FAR-1 and FAR-2 (r = 0.36) and KAN-1 and KAN-3 (r = 0.33).
Fig 5. Biplot of identified helpful bacterial species using Principal Component Analysis (PCA) between two principal component PC1 and PC2 of the river Ganga and Yamuna.
Discussion
The study found that, the river Ganga and Yamuna host several beneficial bacterial genera with enormous taxonomical diversities. Altogether the study could identify 69 beneficial species belonging to 18 genera (Table 2). All the identified beneficial bacteria with their proposed mechanism of action are represented in S1 and S2 Tables. The bacterial communities and their functional genomics in sediments and water of the Apies River, South Africa were analyzed using Metagenomic data. Higher diversity in the microbial species associated with the different land uses in the water and sediments of the Apies River was revealed in this study [21]. The taxonomic classification was also previously used to classify microbe strains with consistent categorization at the species level with appropriate safety evaluation, quality assurance, and non-fraudulent labeling [15, 22–26]. In the present study, the beneficial bacterial species under genus Lactobacillus (L. curvatus, L. brevis, L. helveticus, L. gasseri, L. crisptus and L. casei, etc.) were identified. These Lactobacillus species were reported to exert their beneficial effects by reducing soreness in inflammatory bowel disease (IBD) by producing anti-inflammatory cytokine [27], antibiotic and bacteriostatic activity by the production of bacteriocins [28], and anti-stress activity by the production of β-galactosidase enzymes [29]. L. curvatus, was reported to lower the cholesterol level through enhancement of esterase, lipase, cysteine arylamidase, and β-galactosidase activities in the host organisms [30]. Vibrio spp. were reported to cause health benefits to the host organism by improving disease resistance through the production of bacteriocin-like substance [31], alteration in the hepatosomatic index, and haemocytes number [32]. Bacillus spp. found in the present study were reported to enhance growth, survivability and disease resistance of Labeo rohita, and Macrobrachium rosenbergii etc. through increased alkaline phosphatase activity, globulin content and lysozyme level [33], enhancement of serum lysozyme activity and serum IgM level [34], increased LYZ gene expression [35], etc. The identified Bifidobacterium spp., (B. animalis, B. bifidum, B. longum, B. breve and B. adolescentis, etc.) were reported to attenuate autoimmune encephalomyelitis by inhibiting mononuclear infiltration into the central nervous system [36], diminish gastrointestinal distress by stimulating the production of gastric mucin and other gastrointestinal or neuropeptide hormones [37], anti-obese activities by inhibition of lipid deposit in the liver and adipose tissues [38], alleviate of high-fat diet-induced colitis by inhibition of NF-κB activation and lipopolysaccharide production by gut microbiota [39]. Similarly, Pediococcus spp. was reported to cause many health benefits viz. P. acidilactici was reported to advance reproductive performance [40], P. pentosaceus has anti-inflammation and anti-cancer effects through mitigation of azoxymethane-induced toxicity [41], P. ethanol idurans enhances health through the production of high levels of cellular antioxidant and amplified bile salt hydrolase activities [42]. The identified Enterococcus faecalis, was reported to enhance anti-oxidative activity and anti-tumor activity by NK cells and TNF-α [42]. E. raffinosus which was reported to prevent bacterial infection in Labeo rohita and Labeo catla from E. coli, A. hydrophilla, S. aerous, S. typhimurium [43]. The identified E. hirae, reported producing lipase and bile salt hydrolase enzyme with antioxidant properties, and E. mundtii reported with antimicrobial activity [44]. Four Roseobacter spp., identified from the sediment metagenomes, were reported with therapeutic value for commercial aquaculture. Earlier, several Roseobacter sp. were also reported to reduce fish pathogenic bacteria V. anguillarum by R. clade [45].
The phylogenetic tree analysis showed that the majority of the species are evolutionary diverse. The phylogenetic tree of all the identified beneficial bacteria species was shaped in five different clusters. In CLUSTER-3, L. fermentum and L. helveticus derived from Kanpur and Yamuna sediment samples respectively were found phylogenetically related with a high bootstrap value of 71. A similar observation was reported from Lactobacillus spp. isolated from animal faeces and it was found that, L. salivarius phylogenetic group was closely related to L. animalis, L. apodemi, and L. Murinus [46]. The present finding could be corroborated with a previous report where Lactococcus and Streptococcus appeared to be closely related and Lactobacillus was found to be phylogenetically diverse [27]. The intermixing of phylogenetic distribution, as observed from our study, was also reported previously where Lactobacillus and Pediococcus were phylogenetically intermixed with 5 species of Pediococcus [47]. The Lactobacillus chromosomes also expressed the high heterogeneity at phylogenetic, phenotypic, and ecological levels amid the different members of this genus [48]. The present study also found heterogeneity of clustering in Lactobacillus species and other beneficial bacteria.
Relative abundance study showed that beneficial bacteria species of different genera were variedly distributed among the three locations; few species are highly dominant in one location over others, viz. Pediococcus acidilactici was highly abundant in Kanpur location of river Ganga as compared to other locations. The PCA analysis also showed that the sites at Farraka are closely associated and sites at Kanpur and New Delhi are diverse about the relative abundance of beneficial bacteria. This location-specific change of microbial diversity in the river sediments might be due to differential physiochemical properties and pollution level of the collected sediments. The primary reason for this difference might be due to the release of heavy organic loads and toxic substances (heavy metals, hazardous chemicals, etc.) in some of the selected locations (Kanpur and New Delhi) of these riverine ecosystems through the release of untreated sewage and industrial wastes. It was reported that Kanpur stretch of river Ganga is highly polluted by the untreated effluents from hundreds of tannery industries present in the river bank [49–50]. Very high quantities of diverse heavy metals like Cr, Cu, Pb, Ni, Zn, etc. were found extensively in the water and in the sediments of river Ganga in Kanpur, where pesticide residue like α-HCH, γ-HCH, Dieldrin and Malathion were also reported with a concentration range from 0.190±0.02 to 2.61±0.05 μg/L2 [51]. However, the Farakka stretch of the river Ganga was reported to be less polluted [52]. Like Kanpur stretch of river Ganga, the New Delhi stretch of the river Yamuna was also reported to be severely polluted by heavy metal pollutions due to the release of untreated metropolitan swages, factory effluents, etc. [53, 54]. Therefore, we presume it might be the reason for differences in the relative abundance of beneficial bacteria species among different locations in the river Ganga and Yamuna. Our results could be supported by the previous finding, where the proportion of beneficial microbes in the gastrointestinal microbiota of Bufo raddei was altered due to heavy-metal pollution [55]. This is the first report on the identification of beneficial bacteria in the sediments of the river Ganga and Yamuna, using a metagenomic approach. This study revealed extensive insights on the abundance of native important beneficial microorganisms in these rivers and their functional properties.
Conclusion
Our research indicates that the sediment metagenome of the river Ganga and Yamuna manifests the enriched microbial distribution of beneficial bacteria. The phylogenetic study of identified useful microbial species revealed that the majority of the species are evolutionarily diverse. This study also refers to the clear distinction in the relative abundance of different beneficial bacteria across the sampling sites. Isolation of different beneficial bacteria from these riverine ecosystems would be highly useful for industrial applications in the future.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
Authors are thankful to Mr. Asim Kumar Jana, Senior Technical Assistant, ICAR-CIFRI, Barrackpore, Kolkata, India for sampling and technical assistance. This work has been carried under CABin Scheme.
Data Availability
The metagenomic sequences used in this study have been submitted to the NCBI-SRA database under Accession Nos: SRP190174, SRP190175, SRP189880, SRP191076, SRP191079, SRP191075, SRP191073, SRP191080, and SRP191499.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Zorriehzahra MJ, Delshad ST, Adel M, Tiwari R, Karthik K, Dhama K, et al. Probiotics as beneficial microbes in aquaculture: an update on their multiple modes of action: a review. Veterinary Quarterly. 2016; 36(4):228–241. 10.1080/01652176.2016.1172132 [DOI] [PubMed] [Google Scholar]
- 2.Sánchez B, Delgado S, Blanco‐Míguez A, Lourenço A, Gueimonde M, Margolles A. Probiotics, gut microbiota, and their influence on host health and disease. Molecular nutrition & food research. 2017; 61(1):1600240. [DOI] [PubMed] [Google Scholar]
- 3.Ouwehand A, Isolauri E, Salminen S. The role of the intestinal microflora for the development of the immune system in early childhood. European journal of nutrition. 2002; 41(1):i32–i37. [DOI] [PubMed] [Google Scholar]
- 4.O'sullivan GC. Probiotics. British Journal of Surgery. 2001; 88(2):161–162. 10.1046/j.1365-2168.2001.01656.x [DOI] [PubMed] [Google Scholar]
- 5.Villamil LA, Tafalla C, Figueras A, Novoa B. Evaluation of immunomodulatory effects of lactic acid bacteria in turbot (Scophthalmus maximus). Clin. Diagn. Lab. Immunol. 2002; 9(6):1318–1323. 10.1128/cdli.9.6.1318-1323.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagpal R, Wang S, Ahmadi S, Hayes J, Gagliano J, Subashchandrabose S, et al. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Scientific reports. 2018; 8(1):12649 10.1038/s41598-018-30114-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashraf S, Zaneb H, Yousaf MS, Ijaz A, Sohail MU, Muti S, et al. Effect of dietary supplementation of prebiotics and probiotics on intestinal microarchitecture in broilers reared under cyclic heat stress. Journal of animal physiology and animal nutrition. 2013; 97:68–73. 10.1111/jpn.12041 [DOI] [PubMed] [Google Scholar]
- 8.Rao V. Bioremediation to restore the health of aquaculture pond ecosystem. Advances in Aquatic Ecology. 2007; 1:85–94. [Google Scholar]
- 9.Ramesh D, Souissi S, Ahamed TS. Effects of the potential probiotics Bacillus aerophilus KADR3 in inducing immunity and disease resistance in Labeo rohita. Fish & shellfish immunology. 2017; 70:408–415. [DOI] [PubMed] [Google Scholar]
- 10.Nandi A, Banerjee G, Dan SK, Ghosh K, Ray AK. Evaluation of in vivo probiotic efficiency of Bacillus amyloliquefaciens in Labeo rohita challenged by pathogenic strain of Aeromonas hydrophila MTCC 1739. Probiotics and antimicrobial proteins. 2018; 10(2):391–398. 10.1007/s12602-017-9310-x [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Porchas M, Villalpando-Canchola E, Suarez LE, Vargas-Albores F. How conserved are the conserved 16S-rRNA regions? PeerJ. 2017; 5:e3036 10.7717/peerj.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, et al. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Scientific reports. 2016; 6:33721 10.1038/srep33721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji B, Nielsen J. From next-generation sequencing to systematic modeling of the gut microbiome. Frontiers in genetics. 2015; 6:219 10.3389/fgene.2015.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong J, Xia J. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites. 2017; 7(4):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature communications. 2016; 7:11257 10.1038/ncomms11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987; 4(4):406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences. 2004; 101(30):11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular biology and evolution. 2013; 30(12):2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imchen M, Kumavath R, Barh D, Vaz A, Góes-Neto A, Tiwari S, et al. Comparative mangrove metagenome reveals global prevalence of heavy metals and antibiotic resistome across different ecosystems. Scientific reports. 2018; 8(1):1–15. 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howe E, Holton K, Nair S, Schlauch D, Sinha R, Quackenbush J. Mev: multiexperiment viewer In Biomedical informatics for cancer research 2010. (pp. 267–277). Springer, Boston, MA. [Google Scholar]
- 21.Abia AL, Alisoltani A, Keshri J, Ubomba-Jaswa E. Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies River, South Africa, as a function of land use. Science of the Total Environment. 2018; 616:326–334. 10.1016/j.scitotenv.2017.10.322 [DOI] [PubMed] [Google Scholar]
- 22.Klein G, Pack A, Bonaparte C, Reuter G. Taxonomy and physiology of probiotic lactic acid bacteria. International journal of food microbiology. 1998; 41(2):103–125. 10.1016/s0168-1605(98)00049-x [DOI] [PubMed] [Google Scholar]
- 23.Joint FA. WHO working group report on drafting guidelines for the evaluation of probiotics in food. London, Ontario, Canada. 2002; 30.
- 24.Vankerckhoven V, Huys G, Vancanneyt M, Vael C, Klare I, Romond MB, et al. Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trends in Food Science & Technology. 2008; 19(2):102–114. [Google Scholar]
- 25.Pot B, Coenye T, Kersters K. The taxonomy of microorganisms used as probiotics with special focus on enterococci, lactococci, and lactobacilli. Microecology and Therapy. 1997:11–26. [Google Scholar]
- 26.Holzapfel WH, Haberer P, Geisen R, Björkroth J, Schillinger U. Taxonomy and important features of probiotic microorganisms in food and nutrition. The American journal of clinical nutrition. 2001; 73(2):365s–373s. [DOI] [PubMed] [Google Scholar]
- 27.Park JS, Choi JW, Jhun J, Kwon JY, Lee BI, Yang CW, et al. Lactobacillus acidophilus improves intestinal inflammation in an acute colitis mouse model by regulation of Th17 and Treg cell balance and fibrosis development. Journal of medicinal food. 2018; 21(3):215–224. 10.1089/jmf.2017.3990 [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Guo Y, Chen H, Wei H, Wan C. Potential of Lactobacillus plantarum ZDY2013 and Bifidobacterium bifidum WBIN03 in relieving colitis by gut microbiota, immune, and anti-oxidative stress. Canadian journal of microbiology. 2018; 64(5):327–337. 10.1139/cjm-2017-0716 [DOI] [PubMed] [Google Scholar]
- 29.de Souza BM, Borgonovi TF, Casarotti SN, Todorov SD, Penna AL. Lactobacillus casei and Lactobacillus fermentum strains isolated from mozzarella cheese: probiotic potential, safety, acidifying kinetic parameters and viability under gastrointestinal tract conditions. Probiotics and antimicrobial proteins. 2019; 11(2):382–396. 10.1007/s12602-018-9406-y [DOI] [PubMed] [Google Scholar]
- 30.Zommiti M, Connil N, Hamida JB, Ferchichi M. Probiotic characteristics of Lactobacillus curvatus DN317, a strain isolated from chicken ceca. Probiotics and antimicrobial proteins. 2017; 9(4):415–424. 10.1007/s12602-017-9301-y [DOI] [PubMed] [Google Scholar]
- 31.Carraturo A, Raieta K, Ottaviani D, Russo GL. Inhibition of Vibrio parahaemolyticus by a bacteriocin‐like inhibitory substance (BLIS) produced by Vibrio mediterranei 1. Journal of applied microbiology. 2006; 101(1):234–241. 10.1111/j.1365-2672.2006.02909.x [DOI] [PubMed] [Google Scholar]
- 32.Thompson J, Gregory S, Plummer S, Shields RJ, Rowley AF. An in vitro and in vivo assessment of the potential of Vibrio spp. as probiotics for the Pacific White shrimp, Litopenaeus vannamei. Journal of applied microbiology. 2010; 109(4):1177–1187. [DOI] [PubMed] [Google Scholar]
- 33.Sumathi C, Dillibabu V, Madhuri DK, Priya DM, Nagalakshmi C, Sekaran G. Dietary inclusion of protease producing novel Pontibacter spp. and Bacillus megaterium as a probiotic enhances immune responses in Labeo rohita. Pakistan Journal of Biological Sciences. 2014; 17(4):451 10.3923/pjbs.2014.451.461 [DOI] [PubMed] [Google Scholar]
- 34.Sun YZ, Yang HL, Ma RL, Lin WY. Probiotic applications of two dominant gut Bacillus strains with antagonistic activity improved the growth performance and immune responses of grouper Epinephelus coioides. Fish & shellfish immunology. 2010; 29(5):803–809. [DOI] [PubMed] [Google Scholar]
- 35.Zhen W, Shao Y, Gong X, Wu Y, Geng Y, Wang Z, et al. Effect of dietary Bacillus coagulans supplementation on growth performance and immune responses of broiler chickens challenged by Salmonella enteritidis. Poultry science. 2018; 97(8):2654–2666. 10.3382/ps/pey119 [DOI] [PubMed] [Google Scholar]
- 36.Salehipour Z, Haghmorad D, Sankian M, Rastin M, Nosratabadi R, Dallal MM, et al. Bifidobacterium animalis in combination with human origin of Lactobacillus plantarum ameliorate neuroinflammation in experimental model of multiple sclerosis by altering CD4+ T cell subset balance. Biomedicine & Pharmacotherapy. 2017; 95:1535–1548. [DOI] [PubMed] [Google Scholar]
- 37.Gomi A, Yamaji K, Watanabe O, Yoshioka M, Miyazaki K, Iwama Y, et al. Bifidobacterium bifidum YIT 10347 fermented milk exerts beneficial effects on gastrointestinal discomfort and symptoms in healthy adults: A double-blind, randomized, placebo-controlled study. Journal of dairy science. 2018; 101(6):4830–4841. 10.3168/jds.2017-13803 [DOI] [PubMed] [Google Scholar]
- 38.Kang D, Li Z, Ji GE. Anti-Obesity Effects of a Mixture of Fermented Ginseng, Bifidobacterium longum BORI, and Lactobacillus paracasei CH88 in High-Fat Diet-Fed Mice. Journal of microbiology and biotechnology. 2018; 28(5):688–696. 10.4014/jmb.1801.01016 [DOI] [PubMed] [Google Scholar]
- 39.Lim SM, Kim DH. Bifidobacterium adolescentis IM38 ameliorates high-fat diet–induced colitis in mice by inhibiting NF-κB activation and lipopolysaccharide production by gut microbiota. Nutrition research. 2017; 41:86–96. [DOI] [PubMed] [Google Scholar]
- 40.Mehdinejad N, Imanpour MR, Jafari V. Combined or Individual Effects of Dietary Probiotic Pedicoccus acidilactici and Nucleotide on Growth Performance, Intestinal Microbiota, Hemato-biochemical Parameters, and Innate Immune Response in Goldfish (Carassius auratus). Probiotics and antimicrobial proteins. 2018; 10(3):558–565. 10.1007/s12602-017-9297-3 [DOI] [PubMed] [Google Scholar]
- 41.Dubey V, Ghosh AR, Bishayee K, Khuda-Bukhsh AR. Probiotic Pediococcus pentosaceus strain GS4 alleviates azoxymethane-induced toxicity in mice. Nutrition research. 2015; 35(10):921–929. 10.1016/j.nutres.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 42.Xu S, Liu T, Radji CA, Yang J, Chen L. Isolation, identification, and evaluation of new lactic acid bacteria strains with both cellular antioxidant and bile salt hydrolase activities in vitro. Journal of food protection. 2016; 79(11):1919–1928. 10.4315/0362-028X.JFP-16-096 [DOI] [PubMed] [Google Scholar]
- 43.Sahoo TK, Jena PK, Nagar N, Patel AK, Seshadri S. In vitro evaluation of probiotic properties of lactic acid bacteria from the gut of Labeo rohita and Catla catla. Probiotics and antimicrobial proteins. 2015; 7(2):126–136. 10.1007/s12602-015-9184-8 [DOI] [PubMed] [Google Scholar]
- 44.Grau T, Vilcinskas A, Joop G. Probiotic Enterococcus mundtii isolate protects the model insect Tribolium castaneum against Bacillus thuringiensis. Frontiers in microbiology. 2017; 8:1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharifah EN, Eguchi M. The phytoplankton Nannochloropsis oculata enhances the ability of Roseobacter clade bacteria to inhibit the growth of fish pathogen Vibrio anguillarum. PLoS One. 2011; 6(10):e26756 10.1371/journal.pone.0026756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endo A, Irisawa T, Futagawa-Endo Y, Salminen S, Ohkuma M, Dicks L. Lactobacillus faecis sp. nov., isolated from animal faeces. International journal of systematic and evolutionary microbiology. 2013; 63(12):4502–4507. [DOI] [PubMed] [Google Scholar]
- 47.Ventura M, O'flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, Van Sinderen D, et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nature Reviews Microbiology. 2009; 7(1):61 10.1038/nrmicro2047 [DOI] [PubMed] [Google Scholar]
- 48.Claesson MJ, van Sinderen D, O'Toole PW. Lactobacillus phylogenomics–towards a reclassification of the genus. International Journal of Systematic and Evolutionary Microbiology. 2008; 58(12):2945–2954. [DOI] [PubMed] [Google Scholar]
- 49.Sankararamakrishnan N, Sharma AK, Sanghi R. Organochlorine and organophosphorous pesticide residues in ground water and surface waters of Kanpur, Uttar Pradesh, India. Environment International. 2005; 31(1):113–120. 10.1016/j.envint.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 50.Gowd SS, Reddy MR, Govil PK. Assessment of heavy metal contamination in soils at Jajmau (Kanpur) and Unnao industrial areas of the Ganga Plain, Uttar Pradesh, India. Journal of Hazardous Materials. 2010; 174(1–3):113–121. 10.1016/j.jhazmat.2009.09.024 [DOI] [PubMed] [Google Scholar]
- 51.Beg KR, Ali S. Chemical contaminants and toxicity of Ganga river sediment from up and down stream area at Kanpur. American Journal of Environmental Sciences. 2008; 4(4):362. [Google Scholar]
- 52.Samanta S. Metal and pesticide pollution scenario in Ganga River system. Aquatic ecosystem health & management. 2013; 16(4):454–464. [Google Scholar]
- 53.Sehgal M, Garg A, Suresh R, Dagar P. Heavy metal contamination in the Delhi segment of Yamuna basin. Environmental monitoring and assessment. 2012; 184(2):1181–1196. 10.1007/s10661-011-2031-9 [DOI] [PubMed] [Google Scholar]
- 54.Malik A, Tauler R. Extension and application of multivariate curve resolution-alternating least squares to four-way quadrilinear data-obtained in the investigation of pollution patterns on Yamuna River, India—a case study. Analytica chimica acta. 2013; 794:20–28. 10.1016/j.aca.2013.07.047 [DOI] [PubMed] [Google Scholar]
- 55.Zhang W, Guo R, Yang Y, Ding J, Zhang Y. Long-term effect of heavy-metal pollution on diversity of gastrointestinal microbial community of Buforaddei. Toxicology letters. 2016; 258:192–197. 10.1016/j.toxlet.2016.07.003 [DOI] [PubMed] [Google Scholar]