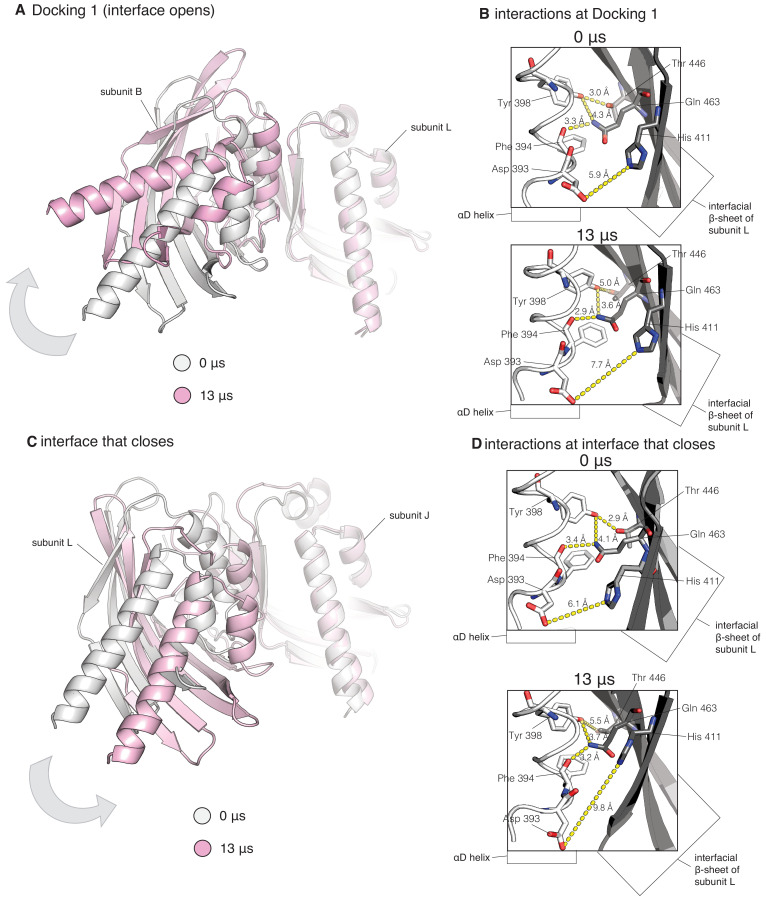
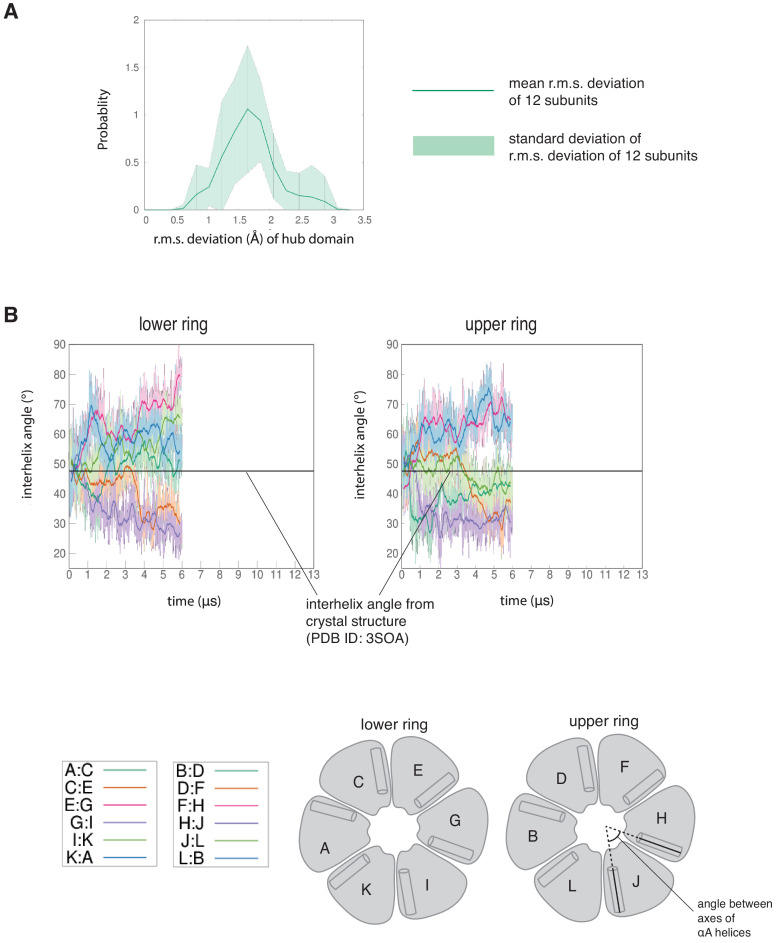
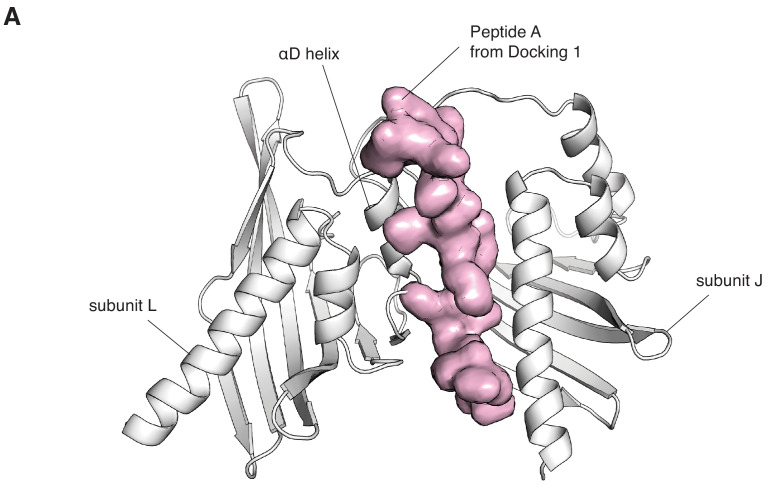
Figure 6. Interfaces at which docking occur are stabilized in an open form, while remaining interfaces close.
(A) Schematic showing the naming scheme used to identify subunits. The interhelix angle between axes of αA helices are used to measure the rotation of each hub domain with respect to the adjacent hub domain. (B) Variation of the angle between the axes of the αA helices of adjacent subunits in the simulation with the regulatory segment. The darker traces are the time-averaged values of the interhelix angles calculated using a moving window of 240 ns, while the lighter shades are the actual distances. The two interfaces where regulatory segments dock between subunits L and B (colored blue), and between subunits F and H (colored pink) open and the remaining interfaces close. In the lower ring the interfaces mirror the behavior of the interfaces in the upper ring. The interhelix angle between the αA helices of adjacent subunits from the crystal structure of an autoinhibited, dodecameric holoenzyme (PDB ID - 3SOA) (Chao et al., 2011), which is perfectly symmetrical, is indicated by the horizontal black line. (C) Variation in the angle between the axes of the αA helices of hub domains that flank the interfaces where the calmodulin-binding elements dock (shown in red), and the distance between a residue from the calmodulin-binding element and the interfacial β-sheet (shown in blue), over the course of the simulation. The darker traces are the time-averaged values, calculated using a moving window of 12 ns, while the lighter shades are the actual distances. The interhelix angle at the start of the simulation is indicated by a horizontal grey line.