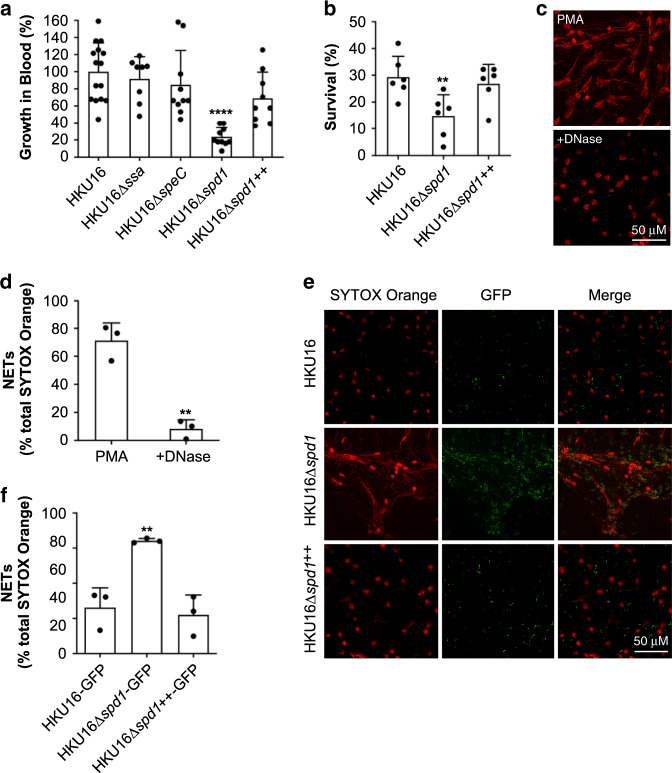
Fig. 3. The ΦHKU.vir-encoded DNase Spd1 promotes resistance to neutrophil killing.
a Growth of indicated HKU16 strains in whole human blood. Statistical significance was assessed using one-way ANOVA with Dunnett’s multiple comparisons post hoc test against the HKU16 control group (****p < 0.0001 for HKU16∆spd1). b Human neutrophil killing assay. The data represent the mean ± SEM of six independent experiments. Statistical significance was assessed using one-way ANOVA with Dunnett’s multiple comparisons post hoc test against the HKU16 control group (**p = 0.0093 for HKU16∆spd1). c Purified human neutrophils were stimulated with 25 nM PMA for 3 h to induce neutrophil extracellular traps (NETs). NETs were detected using the extracellular DNA stain SYTOX Orange (red) and images captured using confocal microscopy. Panels show formation of NETs (left) and NET degradation following incubation with bovine pancreatic DNase I as a positive control (right). d NET quantification of PMA-stimulated neutrophils in the absence or presence of DNase I. Statistical significance was assessed by two-tailed unpaired Student’s t test (**p = 0.0016 for DNase treatment). e Representative images of PMA-stimulated neutrophils following infection with GFP fluorescent GAS (green) for 30 min at a multiplicity of infection of 10 (bacterial CFU:neutrophil). Scale bars represent 50 μm. f NET quantification of PMA-stimulated neutrophils following incubation with GAS. NET quantification is expressed as a percentage of total SYTOX Orange stained area calculated from a minimum of five randomly selected microscopic fields. Error bars represent the mean ± SEM from three independent experiments. Statistical significance was assessed using one-way ANOVA with Dunnett’s multiple comparisons post hoc test against the HKU16-GFP control group (**p = 0.0041 for HKU16∆spd1-GFP). Source data are provided as a Source Data file.