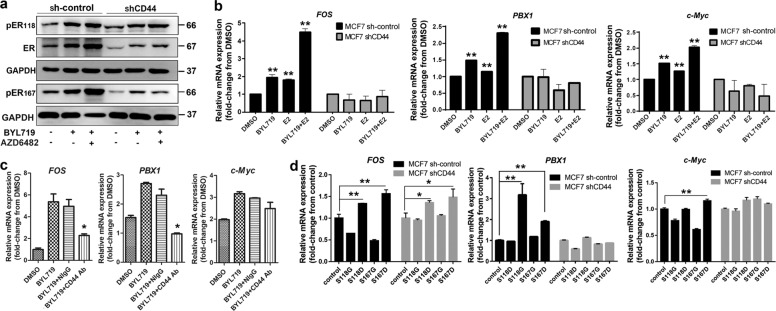
Fig. 3. CD44 mediates ER-dependent transcription upon PI3Kα pathway inhibition.
a Knockdown of CD44 expression attenuated ER activation triggered by PI3K inhibitors. Expression of ER and phosphorylated ER (Ser118 and Ser167) in control or sh-CD44-transfected MCF7 cells depleted of hormones for 3 days and subsequently treated with DMSO, BYL719 (1 μM), or combination BYL719 and Azd6482 (1 μM) treatment for 3 days was measured by immunoblotting. b Expression of candidate target genes in control or sh-CD44-transfected MCF7 cells depleted of hormones for 3 days treated with DMSO, E2 (100 nM), BYL719 (1 μM), or BYL719 combined with BYL719 (1 μM) and E2 (100 nM) for 24 h was measured by RT-qPCR. Values are presented as the mean (±SD). The P-values were calculated using Student’s t-test. All data represent at least three biological repeats. *P < 0.05, *P < 0.01 vs. control. c Expression of candidate target genes in cells depleted of hormones for 3 days and treated with DMSO, BYL719 (1 μM), anti-CD44 antibody (25 μg/ml), or both BYL719 (1 μM) and CD44 antibody (25 μg/ml) for 24 h was measured by RT-qPCR. Normal mouse IgG (NIgG) was used as negative control. Values are presented as the mean (±SD). Results are expressed as mean (±SD), n = 3. *P < 0.05 when compared to BYL719 treatment with or without NIgG. d Effects of phosphomimetic and phospho-dead ER on the transcription of ER target genes in naïve MCF7 or sh-CD44 MCF7 cells. Exogenously phosphomimetic and phospho-dead ER (S118 and ER S167) were prepared. Two codons of ESR1 gene encoding for serine (S) residues were mutated to encode either an aspartic acid (D) residue (ER118S→D, ER167S→D) or glycine (G) residue (ER118S→G, ER167S→G). The phosphomimetic (S118D and S167D) and phospho-dead (S118G and S167G) were expressed in cells by lentivirus transfection. The expression of ER target genes was then measured by RT-qPCR. An empty vector was used as a negative control (vector). Values are presented as the mean (±SD). All data represent at least three biological repeats. *P < 0.05, *P < 0.01.