Figure 2.
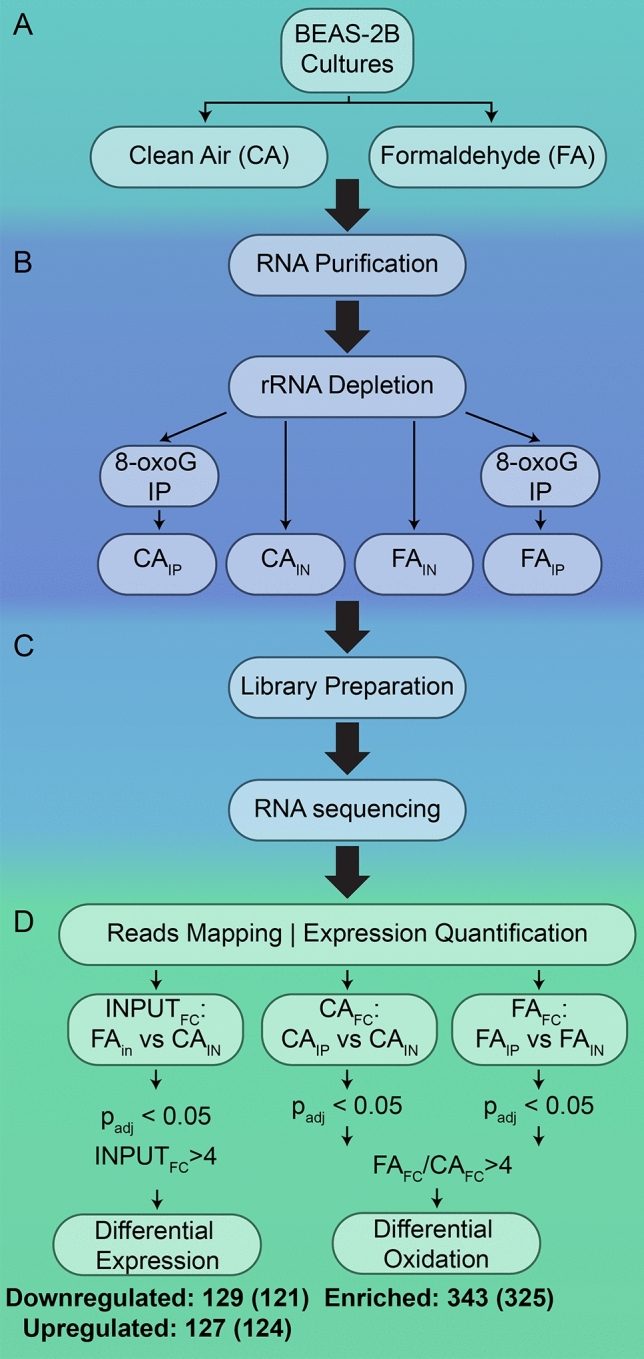
Schematic of 8-oxoG RIP-seq experimental workflow. (A) BEAS-2B cell cultures were exposed to 1 ppm formaldehyde or clean air. (B) Total RNA was extracted and ribosomal RNA (rRNA) was selectively depleted to yield a pool of enriched whole transcriptome RNA. A fraction of this pool was mixed with an anti-8-oxo-7,8-dihydroguanosine (8-oxoG) antibody followed by protein A magnetic beads. The antibody-bound RNA was recovered by competitive elution with excess of free 8-oxoG nucleotides. (C) Both pools, the transcriptome pool and the 8-oxoG transcript pool from each condition, were submitted for Illumina RNA sequencing. (D) Transcript sequences were processed and statistically identified. The number of transcripts identified in the analyses are described in the box of the test. Numbers in parentheses indicate the number of unique gene products identified by Enrichr corresponding to the transcript set.
