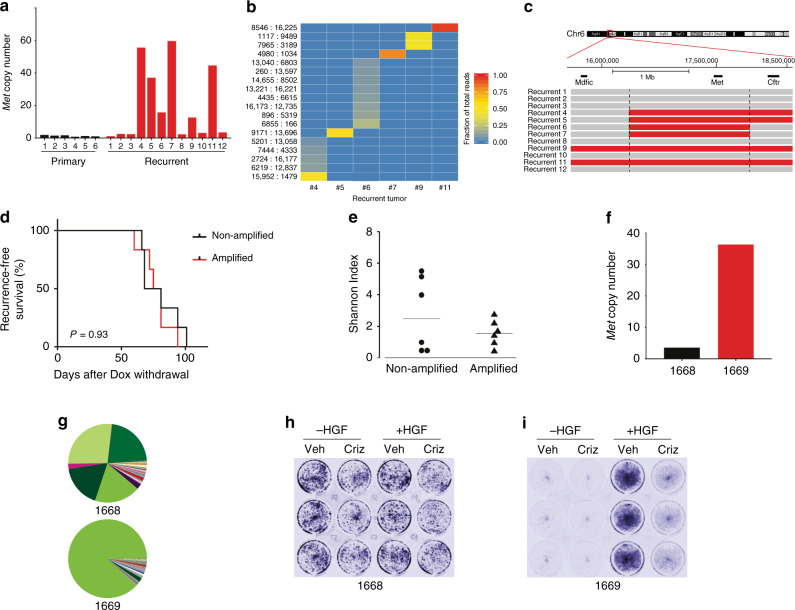
Fig. 4. Met amplification drives recurrence in a subset of clonal recurrent tumors.
a qPCR analysis of Met copy-number in primary and recurrent tumors. Data are expressed as fold-increase in Met copy number relative to blood. n = 6 biologically independent primary tumors and n = 12 biologically independent recurrent tumors. b Heatmap of the abundance of selected barcodes in recurrent tumors, demonstrating that individual Met-amplified tumors have unique barcodes. All barcodes present at >5% in any tumor are shown. c Met-amplified recurrent tumors have distinct Met amplicons. qPCR analysis was used to measure Mdfic, Met, and Cftr copy number in individual Met-amplified tumors. Amplified regions are shown in red. d Kaplan–Meier recurrence-free survival curves for orthotopic tumors, stratified based on Met amplification status. Differences in survival were calculated using the Kaplan–Meier estimator with the log-rank (Mantel-Cox) test. P = 0.637. e Shannon Diversity Index showing barcode complexity in recurrent tumors with and without Met amplification. n = 6 biologically independent recurrent tumors without Met amplification and n = 6 biologically independent recurrent tumors with Met amplification. f Met copy number in two cell lines derived from orthotopic recurrent tumors with or without Met amplification. n = 1 cell line without Met amplification and n = 1 cell line with Met amplification. g Pie charts showing the relative frequency of barcodes in cells from f. h–i Met signaling drives tumor cell proliferation in Met-amplified recurrent tumor cells. Recurrent tumor cells 1668 (h) or 1669 (i) were grown in the presence of HGF and/or crizotinib for 3 days. Cells were stained using crystal violet.