Figure 3.
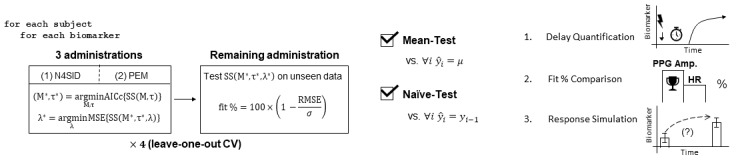
Modeling, optimization, and cross-validation; quality assurance; and analysis. (Left) Modeling, optimization, and testing were performed in a leave-one-out cross-validation (CV) scheme, where, out of the four administrations, each administration was considered once as the unseen test set. The model order (M) and input delay (τ) were optimized by minimizing the small sample size-corrected Akaike information criterion (AICc). For each model order-input delay combination, a state-space model was trained by first initializing the parameter estimates using subspace estimation (N4SID); this was then followed by prediction error minimization (PEM) to refine the parameter estimates. Ridge regression was then performed for this specific model configuration by iterating lambda (λ) logarithmically over a specified interval and minimizing the mean square error. The 1-step-ahead prediction performance was evaluated using a fit percentage formula based on the root mean square error (RMSE) normalized by the standard deviation of the data (σ). (Middle) For quality assurance beyond subjective satisfaction in visual results, the models were evaluated against two objective baseline tests from the literature: the mean test and the naïve test. (Right) To extract the model information pertinent to a deepened dynamic understanding of biomarker responses, (1) optimal input delays were compiled to assess the expected response latency following stimulation onset, (2) the biomarkers were compared against each other to identify superiority in monitoring dynamic changes following transcutaneous cervical vagus nerve stimulation (tcVNS), and (3) the responses were quantifiably visualized via controlled simulation to produce population trajectories of expected dynamic changes following transcutaneous cervical vagus nerve stimulation (tcVNS) administration, in comparison to sham.
