Abstract
Biomechanical contrast within tissues can be assessed based on the resonant frequency probed by spectroscopic magnetomotive optical coherence elastography (MM-OCE). However, to date, in vivo MM-OCE imaging has not been achieved, mainly due to the constraints on imaging speed. Previously, spatially-resolved spectroscopic contrast was achieved in a “multiple-excitation, multiple-acquisition” manner, where seconds of coil cooling time set between consecutive imaging frames lead to total acquisition times of tens of minutes. Here, we demonstrate an improved data acquisition speed by providing a single chirped force excitation prior to magnetomotion imaging with a BM-scan configuration. In addition, elastogram reconstruction was accelerated by exploiting the parallel computing capability of a graphics processing unit (GPU). The accelerated MM-OCE platform achieved data acquisition in 2.9 s and post-processing in 0.6 s for a 2048-frame BM-mode stack. In addition, the elasticity sensing functionality was validated on tissue-mimicking phantoms with high spatial resolution. For the first time, to the best of our knowledge, MM-OCE images were acquired from the skin of a living mouse, demonstrating its feasibility for in vivo imaging.
Biomechanical properties of tissues can be indicative of the underlying pathophysiological conditions and hence have been widely investigated for disease diagnosis [1]. Magnetomotive optical coherence elastography (MM-OCE) utilizes optical coherence tomography (OCT) to sense cross-sectional tissue displacements which are magnetically induced, to thereby assess tissue viscoelasticity [2–7]. MM-OCE allows for remote manipulation of tissue dynamics by using magnetic nanoparticles (MNPs). With a high magnetic susceptibility, MNPs that are deployed to the biological tissues can respond to an external magnetic field with high sensitivity and specificity. In fact, magnetic force can be generated by an alternating magnetic field gradient, which induces “magnetomotion” of MNP-laden tissues. Afterward, the subtle motion can be detected with phase-sensitive OCT with sub-nanometer displacement sensitivity. Spectral analysis of the magnetomotion upon specific force exertion can be further characterized for elasticity assessment. The natural or resonant frequency (f0) can be extracted as it is proportional to the square root of the Young’s modulus (E), i.e., , for a Kelvin-Voigt medium measured in the linear elastic regime [2,3]. Compared to other OCE techniques, MM-OCE does not require precise quantification of force [6], and the relatively low force applied has made it suitable for imaging very soft tissues [1].
As a dynamic OCE approach, MM-OCE can extract biomechanical characteristics via various excitation-detection combinations. Mainly, MM-OCE probes either the transient or spectroscopic mechanical response. For instance, a step-wise force excitation induces an underdamped harmonic oscillation and allows for the quantification of natural frequency and creep parameter, which can subsequently be utilized to infer tissue viscoelasticity [2,5]. In addition, by exerting a chirped force, a spectroscopic mechanical response can be obtained, where the Lorentzian-shaped mechanical spectrum allows for the assessment of the resonant frequency (f0) and the spectral peak width, which are associated with the elasticity and viscosity, respectively [3]. From a signal processing point-of-view, spectroscopic MM-OCE can provide a higher excitation energy, and hence, results in a better signal-to-ratio (SNR) as compared to step-response MM-OCE due to the exertion of the broadband force for a longer period of time. Initially, the concept of spectroscopically revealed mechanical contrast was demonstrated with an external vibration source [8]. Spectroscopic MM-OCE, which provides internal loading via magnetic modulation, has also demonstrated its biomechanics-sensing functionality for various applications, such as visualizing the tumor margin of ex vivo human breast tissue [6], probing stiffness changes in magnetic hyperthermia treated chicken breast [7], and revealing the spectroscopic response of blood clots [4] and ear drum samples [9].
Spectroscopic MM-OCE typically probes the bulk biomechanical response of a presumably homogeneous sample with M-mode data acquisition, which limits the spatial accessibility across the sample. Previously, two-dimensional mechanical contrast was obtained by collecting multiple MM-OCT images that were excited with a wide range of single-tone modulation frequencies (fm), one at a time, followed by the combination of fm-dependent local magnetomotion amplitudes to reconstruct a mechanical spectrum at each spatial location [6]. In fact, spatially-resolved elastic contrast can also be observed from the local magnetomotion, as the closer fm is to f0 (which is elasticity-dependent), the greater the displacement. For each MM-OCT acquisition, a sinusoidal force oscillating at a certain fm was provided for 1–2 s as an oversampled B-mode frame was collected to detect the magnetomotion across a lateral range. To avoid coil overheating and permanent damage, a pause of a few seconds was required between each frame, leading to total acquisition times in the tens of minutes. Therefore, while this “multiple excitation (fm-swept)” technique has shown success in imaging ex vivo heterogenous specimens [6], the overall acquisition time was too long and hindered the possibility for in vivo imaging.
Here, we demonstrate an accelerated MM-OCE platform that allows for rapid, in vivo, two-dimensional spectroscopic MM-OCE imaging. Novel improvements for speedup in both data acquisition and post-processing sessions are shown. For the data acquisition time, the major bottleneck of the previous method was the long inter-frame time interval that was purposely set to prevent coil overheating as alternating currents passed through the coil. To eliminate this time-consuming factor, we directly applied a broadband magnetic force in a chirped waveform and acquired the OCT data in a single BM-mode scanning configuration (i.e., repetitive B-mode acquisition at the same image plane with negligible time delay between frames) with a higher frame rate. Therefore, a significant decrease in the acquisition time for a single MM-OCE measurement resulted. Recently, rapid BM-scanning has drawn increasing attention in dynamic OCE, as demonstrated in shear wave OCE methods [10,11]. A similar scanning scheme was also employed to achieve rapid volumetric MM-OCT that detected the presence of MNPs based on the dynamic contrast. Despite this, the in vivo imaging capability has never been demonstrated [12].
With BM-scan MM-OCE data stacks being collected, it is highly desirable to enable rapid post-processing as well, as real-time visualization allows for better optimization of imaging parameters during the experiment, and greatly benefits in vivo imaging. Graphics processing unit (GPU) chips are well-known for their parallel computing ability, and were utilized to accelerate image reconstruction. With the capability of launching multiple kernels in parallel, GPU chips can operate at 10× memory bandwidth compared to central processing unit (CPU) chips [13]. In fact, GPU-assisted computation has enabled real-time OCT imaging [14], in vivo interferometric synthetic aperture microscopy [15], and near video-rate quasistatic OCE [16]. Here, for the first time, GPU-assisted data processing is implemented for MM-OCE.
To validate the proposed method, MNP-mixed tissue-mimicking phantoms with different elasticity were first fabricated and imaged. The cylindrical samples (∅~39 mm, height ~8 mm) were produced by mixing polydimethylsiloxane fluid (PDMS) with curing agent (RTVA) and crosslinker (RTVB) at mass ratios ranging from 30:10:1 to 200:10:1 (PDMS:RTVA:RTVB). Note that the higher the proportion of PDMS, the softer the sample. Once the mixture solidified after oven baking (80°C, 6–8 h), PDMS phantoms with Young’s moduli (E) ranging from 0.9–90 kPa were produced, where E was measured by conventional mechanical indentation testing and classic Hertzian model fitting. For all samples, 2 mg/g of magnetite MNPs (Sigma #637106) and 1 mg/g of TiO2 were added as mechanical perturbative agents and as optical scatterers, respectively. In addition, a side-by-side heterogenous phantom was also fabricated with a PDMS:RTVA:RTVB ratio of 30:10:1 (stiff) and 200:10:1 (soft). To demonstrate the feasibility for in vivo imaging, the skin at the left flank of a female mouse (C57BL6) was injected with 70 mg/g of dextran-coated maghemite MNPs (Sigma #544884) and imaged before and after CO2 euthanasia. Dextran-coating of the MNPs was performed following [17] to enhance MNP solubility and cytotoxicity. The animal experiment was conducted under a protocol approved by the Institutional Animal Care and Use Committee of the University of Illinois at Urbana-Champaign.
The MM-OCE setup [Fig. 1(a)] used in this study was similar to that described in our previous study [6,7,9,18]. In brief, a magnetic coil was placed above the MNP-laden sample to exert the magnetic force (coil activated with ≤ 8 V). Magnetomotion was detected by a spectral-domain OCT system (superlu-minenscent diode source, 1325±50 nm center wavelength and bandwidth, 1024 pixel InGaAs line-scan camera), with an axial resolution ~8 μm, transverse resolution ~16 μm, and a common-path phase noise between frames of ~2.2 nm. During MM-OCE imaging, a chirped magnetic excitation was provided. M-mode data were acquired for 4096 lines at a ~3 kHz scan rate, and the mechanical spectrum was obtained as previously described [7]. In addition, 1024 or 2048 frames of BM-mode data were collected during a single chirped excitation (10–300 Hz) [Fig. 1(b)]. Each frame scanned ~2 mm laterally with 125 scans acquired at a ~92 kHz line scan rate (1.36 ms per frame). Magnetomotion at each cross-sectional location was extracted from phase differences between consecutive frames, whose Fourier transform (FFT) produced a mechanical spectrum, and where the local f0 values were obtained by allocating the amplitude peaks. Note that the phase differences were calculated based on the complex signals [8]. In addition, subtraction between adjacent lines along the fast axis was also performed as a preprocessing step to remove fixed pattern noise [19]. For most samples, data collection was made both with the magnetic field (Bon) and without (Boff) so that the transfer function could be calculated. For the heterogeneous phantom, additional data processing steps were performed to visualize the cross-sectional magnetomotion amplitudes at a specific fm. In fact, bandpass filtering around fm was performed on individual mechanical spectra, and the envelope of the complex analytical signal at each spatial location was computed afterward. For comparison, the same heterogeneous phantom was also imaged by the multiple excitation (fm-swept) method as previously reported in [6], where fm was given from 5 Hz to 300 Hz with a 5 Hz interval. Each acquisition contained a Bon=Boff pair of oversampled B-frames, where each frame was composed of 4096 scans acquired at a ~3 kHz line rate over ~2 mm. A delay of at least 5 sec occurred between each frame to prevent coil overheating.
Fig. 1.
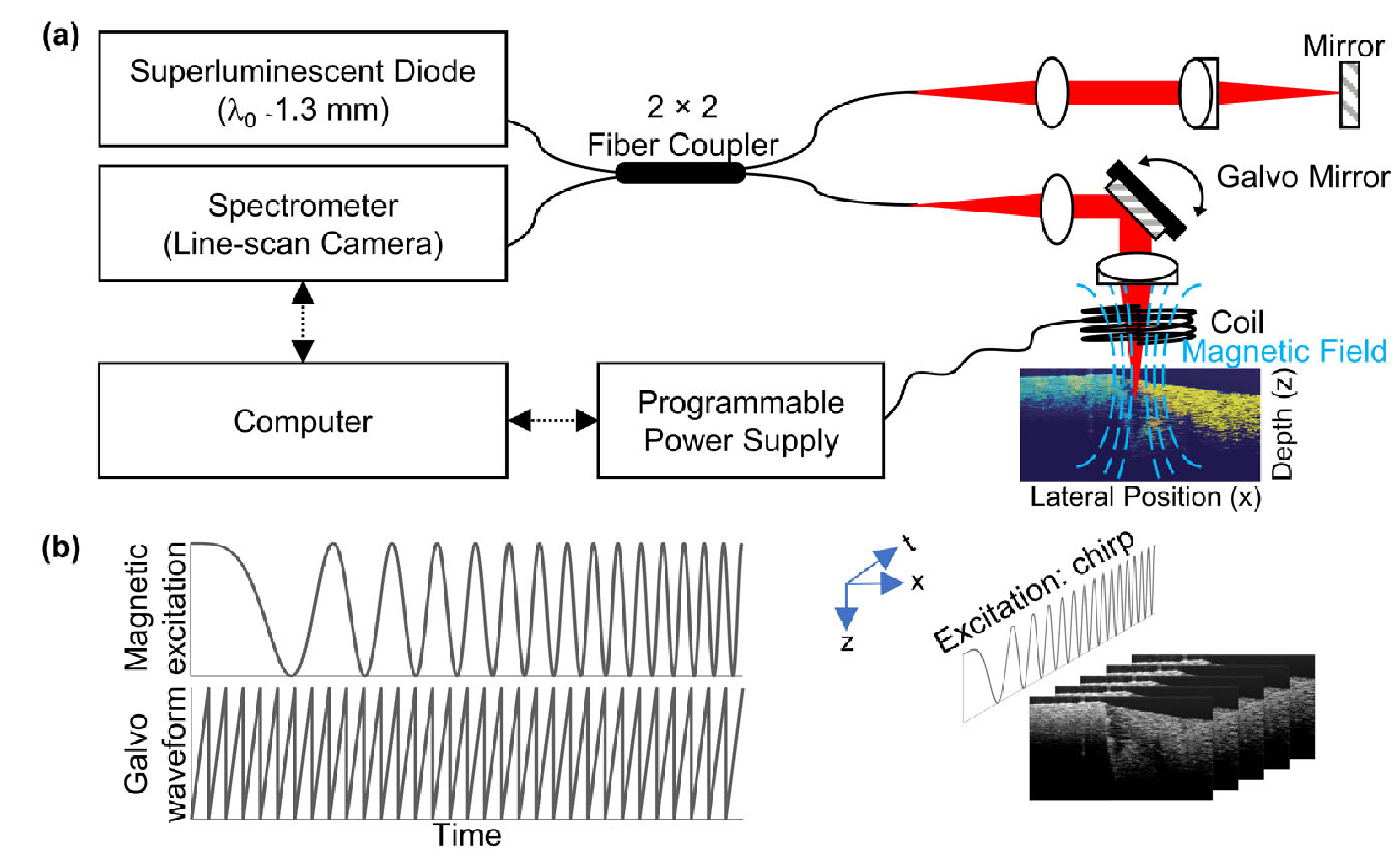
(a) System schematic for magnetomotive optical coherence elastography (MM-OCE). (b) Illustration of the proposed data acquisition acceleration method with a chirped mechanical excitation and BM-mode scanning configuration.
Parallel computing was enabled with a commercially available graphics card (GeForce RTX 2080, Nvidia) to accelerate data processing. The algorithms (Fig. 2) were implemented in a compute unified device architecture platform (CUDA 10.0, Nvidia), where the image processing functions were compiled into a dynamic-link library (DLL) file via Visual Studio 2017 (Microsoft). Subsequently, the DLL functions could be accessed by LabVIEW 2018 (National Instruments), the same interface for data acquisition. To accommodate for high speed data collection, each raw frame was first stored in host buffers before being sequentially transferred to GPU memory, as typically performed [14]. Afterward, standard OCT processing of each frame was executed by a single kernel in GPU, where parallelism among A-scans was exploited. In fact, a large portion of the data would be OCT-processed and stored in GPU memory by the time the full BM-scan stack was acquired. Subsequently, spectral analysis and f0 allocation were carried out along the slow-axis (frames) for all the depths, in parallel. Note that median filtering was performed with ArrayFire API [20]. Shared memory was not employed. The personal computer was equipped with an Intel Core i7 CPU processor and 24 GB of random access memory (RAM). To reduce the computational burden, only the first 256 pixels in depth, where most of the scattering signal was present, was processed in each frame. To validate the results of GPU-assisted processing, CPU-only-based post-processing programmed in Matlab 2016a (Mathworks) was also performed for comparison.
Fig.2.
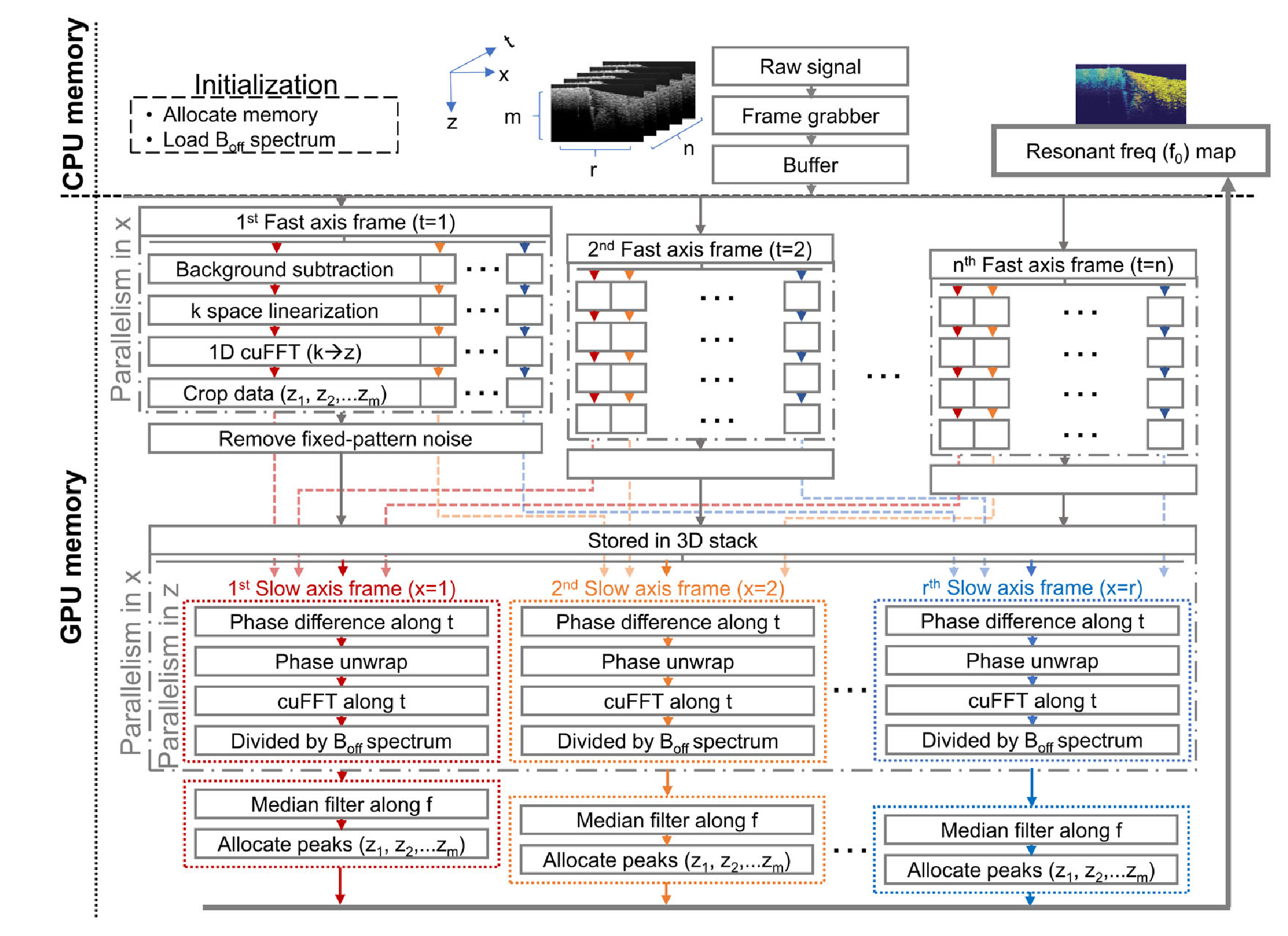
Software architecture for data acquisition and processing.
Cross-sectional elastograms (f0 maps) of the tissue-mimicking PDMS phantoms revealed clear contrast among samples with various stiffness, as shown in Fig. 3. The mechanical spectra obtained from BM-mode data agreed well with the conventional M-mode MM-OCE data. In addition, an expected linearity between the retrieved f0 values and were observed [2,3]. Lastly, the f0 map acquired with and without GPU-assisted processing appeared identical. In fact, the median f0 obtained from the maps with and without GPU exhibited no difference and the standard deviation of f0 has a relative error ≤ 5.67 × 10−2. The empirical results validated the accuracy of the accelerated MM-OCE platform.
Fig. 3.
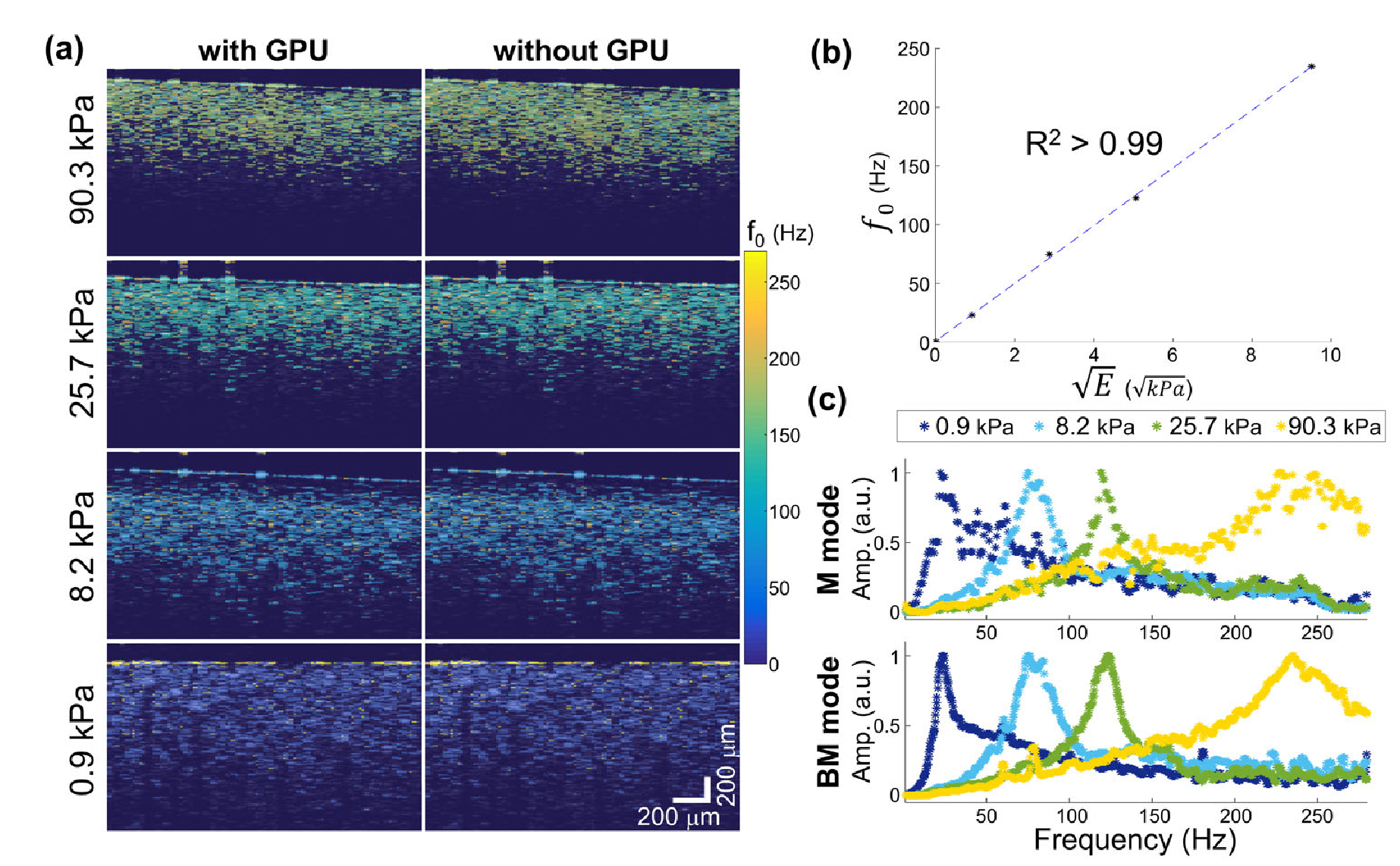
Validation of the accelerated MM-OCE platform demonstrated on homogeneous phantoms. Comparisons are shown between (a) data processing with and without GPU-assistance, between (b) f0 and indentation results, and between (c) M-mode and BM-mode spectra.
The acquisition time (Table 1) for one BM-scan stack (2048 frames) after single chirped magnetic excitation was 2.9 s, which was a speedup of 414×, as compared to the multiple excitation method (~20 min). In addition, the fact that GPU-assisted algorithms were accessible in LabVIEW greatly reduced the data processing time (Table 2), as pre-processing (OCT processing) was initiated immediately after the acquisition of the first fast-axis frame, and post-processing (OCE processing) began once the entire BM-scan stack was acquired. OCT-processing for a frame took as little as ~4.6 ms, and the computation time for f0 allocation and spatial mapping took ~618.8 ms. In contrast, CPU-based processing required a total of 81.3 s to load, process, and render the f0 map. Moreover, without implementing the algorithms in LabVIEW, CPU-based processing could only occur after the entire BM-scan data stack was saved. Therefore, a speedup of 131× was achieved for the post-processing computation. Note that prior to the “Bon” data collection, “Boff” data was first collected, and a Boff Fourier spectrum averaged over the high-SNR area was processed in 65.8 ms on the accelerated platform (as compared to 44.3 s for CPU-based processing).
Table 1.
Data Acquisition Time Comparison
| Ahmad’s Method [6] | Proposed Method | Speedup |
|---|---|---|
| ~20 min | 2.9 s | 414× |
Table 2.
Post-Processing Time Comparison
| n | CPU-Based Processing | GPU-Assisted Processing | Speedup |
|---|---|---|---|
| 2048 | 81.3 s | 618.8 ms | 131× |
| 1024 | 40.1 s | 276.1 ms | 145× |
Shown in the heterogenous phantom (Fig. 4), the spectroscopic response acquired from the proposed scheme was similar to that from the multiple excitation (fm-swept) method. In both cases, stronger displacements were shown on the softer side at a lower fm, versus the stiffer side at a higher fm. This was expected as the maximum displacement occurs at the natural mechanical resonance frequency, and where f0 is positively correlated with stiffness. The f0 difference between the two sides was also seen in the reconstructed mechanical spectra. Interestingly, the proposed BM-scan method manifested a better spatial resolution (pixel-level) as compared to the previous method. The lateral blurs in Fig. 4(a) reflects the global processing nature of the lateral FFT. In contrast, this work [Fig. 4(b)] performs the FFT along frames, which does not involve the use of the lateral FFT along oversampled lateral positions, which are coupled with time. Although the frame rate of this work limits the mechanical spectrum reconstruction at higher frequencies, the fact that the spatial oversampling criteria is relaxed suggests that pixel-resolution elastograms are feasible, especially with the potential incorporation of a MHz Fourier-domain mode-locked OCT system with 16 kHz frame rate [11]. Nevertheless, the temporal resolution in this work (~600 fps) was acceptable as most soft tissues manifest an f0 no greater than 200 Hz [5,7].
Fig. 4.
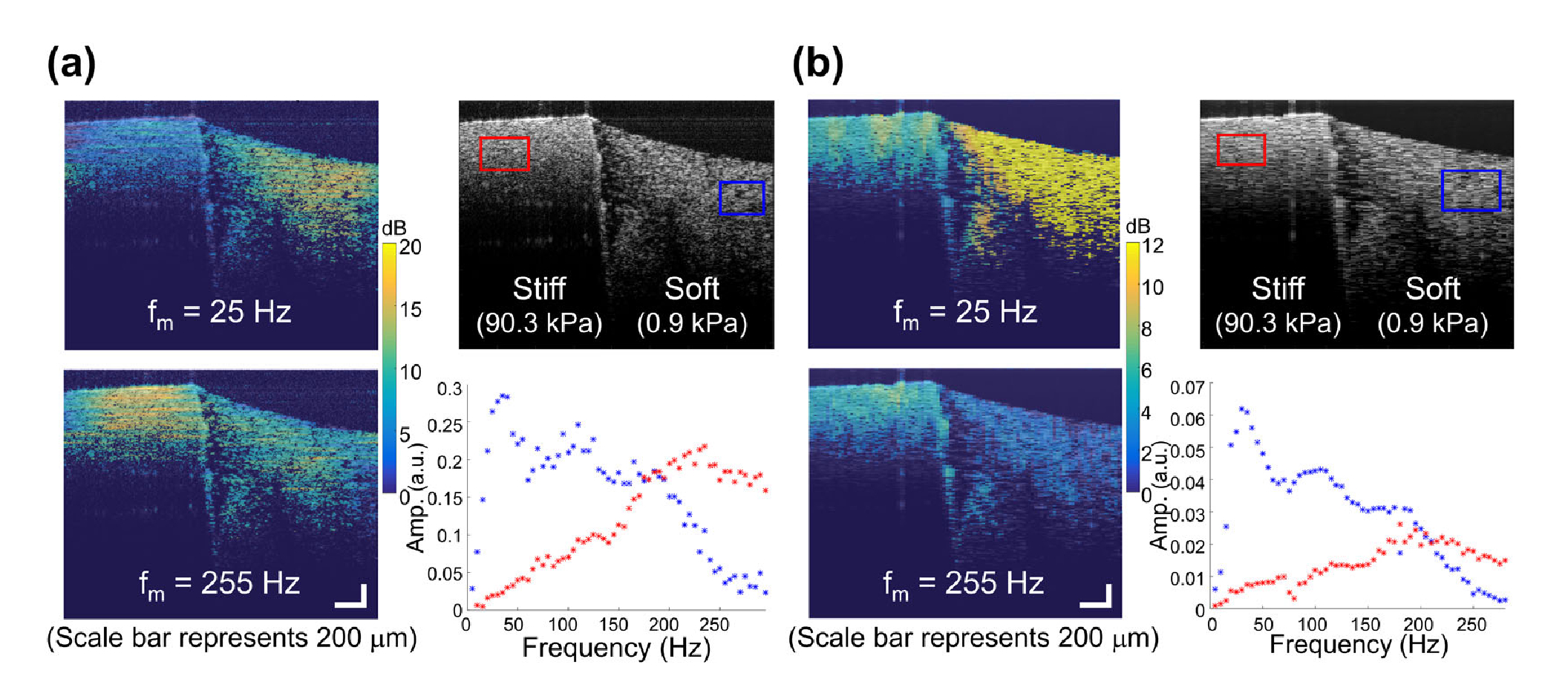
Comparison of (a) the previous method [6] and (b) the current method with demonstration on a heterogeneous phantom. (a) Multiple excitation (fm-swept) (Ahmad’s method [6]). (b) Single chirped excitation (proposed method).
The feasibility of in vivo biological imaging via the accelerated MM-OCE platform is also demonstrated. A total of 1024 frames were collected in ~1.45 s as the chirped magnetic force was applied to the MNP-laden mouse skin, followed by 276.1 ms of post-processing time. In Fig. 5, similarity can be found between the f0 map of the same mouse skin before (in vivo) and immediately after euthanasia (ex vivo). Interestingly, more widely distributed values of f0 are seen at the in vivo elastogram, which was likely due to subtle blood flow motion in the living mouse. Nevertheless, the mean f0 values measured from the in vivo (166.9 Hz) and ex vivo (170.4 Hz) mouse skin were similar. In addition, both values do not deviate dramatically (~20%) from the f0 probed from the M-mode data (140.2 Hz), which was acquired with a 10–800 Hz chirped force excitation.
Fig. 5.
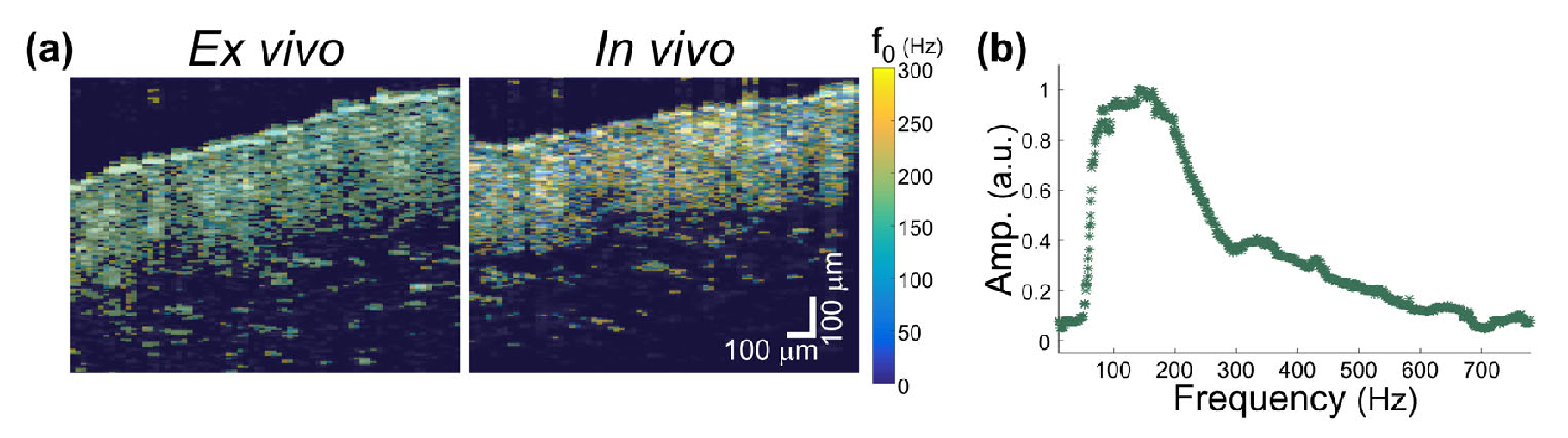
MM-OCE imaging of ex vivo and in vivo mouse skin. (a) f0-based elastograms. (b) M-mode mechanical spectrum.
In summary, we have developed a novel accelerated MM-OCE platform that successfully mapped the mechanical contrast in an in vivo biological tissue for the first time. The single-shot method resulted in data acquisition in ≤ 2.9 s and OCE post-processing in ≤ 618.8 ms, a speedup of at least 414× and 131×, respectively, over previous methods, without sacrificing the mechanical sensing accuracy. Additionally, magnetomotion at each pixel can be obtained without the constraints of spatial oversampling imposed in typical MM-OCT imaging. In the future, a MHz laser source with a resonant scanner [10] can be incorporated in the OCT hardware, and OpenGL-based image rendering can be employed in the software to further improve the imaging speed. Achieved with a single force excitation, the accelerated MM-OCE platform is potentially a powerful tool for in vivo MM-OCE applications, offering new opportunities for bridging the gap between fundamental research and clinical applications in the future.
Funding.
National Institutes of Health (1R01CA213149, 1R01EB013723).
Footnotes
Disclosures. The authors declare no conflicts of interest.
REFERENCES
- 1.Kennedy BF, Kennedy KM, and Sampson DD, IEEE J. Sel. Top. Quantum Electron 20, 272 (2014). [Google Scholar]
- 2.Crecea V, Oldenburg AL, Liang X, Ralston TS, and Boppart SA, Opt. Express 17, 23114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oldenburg AL and Boppart SA, Phys. Med. Biol 55, 1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldenburg AL, Wu G, Spivak D, Tsui F, Wolberg AS, and Fischer TH, IEEE J. Sel. Top. Quantum Electron 18, 1100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crecea V, Ahmad A, and Boppart SA, J. Biomed. Opt 18, 121504 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad A, Huang P-C, Sobh NA, Pande P, Kim J, and Boppart SA, Phys. Med. Biol 60, 6655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang P-C, Pande P, Ahmad A, Marjanovic M, Spillman DR, Odintsov B, and Boppart SA, IEEE J. Sel. Top. Quantum Electron 22, 104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adie SG, Liang X, Kennedy BF, John R, Sampson DD, and Boppart SA, Opt. Express 18, 25519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang P-C, Chaney EJ, Shelton RL, and Boppart SA, IEEE Trans. Biomed. Eng 65, 2837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh M, Wu C, Liu C-H, Li J, Schill A, Nair A, and Larin KV, Opt. Lett 40, 2588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambroziński Ł, Song S, Yoon SJ, Pelivanov I, Li D, Gao L, Shen TT, Wang RK, and O’Donnell M, Sci. Rep 6, 38967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad A, Kim J, Shemonski ND, Marjanovic M, and Boppart SA, J. Biomed. Opt 19, 126001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirk DB and Wen-Mei WH, Programming Massively Parallel Processors: A Hands-On Approach (Morgan Kaufmann, 2016). [Google Scholar]
- 14.Jian Y, Wong K, and Sarunic MV, J. Biomed. Opt 18, 026002 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Ahmad A, Shemonski ND, Adie SG, Kim H-S, Hwu W-MW, Carney PS, and Boppart SA, Nat. Photonics 7, 444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirk RW, Kennedy BF, Sampson DD, and McLaughlin RA, J. Lightwave Technol 33, 3481 (2015). [Google Scholar]
- 17.Yu M, Huang S, Yu KJ, and Clyne AM, Int. J. Mol. Sci 13, 5554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang P-C, Chaney EJ, Iyer RR, Spillman DR, Odintsov B, Sobh NA, and Boppart SA, Biomed. Opt. Express 10, 539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J-H, Han J-H, and Jeong J, Laser Phys. Lett 10, 045604 (2013). [Google Scholar]
- 20.Yalamanchili P, Arshad U, Mohammed Z, Garigipati P, Entschev P, Kloppenborg B, Malcolm J, and Melonakos J, ArrayFire (Atlanta: AccelerEyes, 2015), https://github.com/arrayfire/arrayfire. [Google Scholar]


