Figure 4.
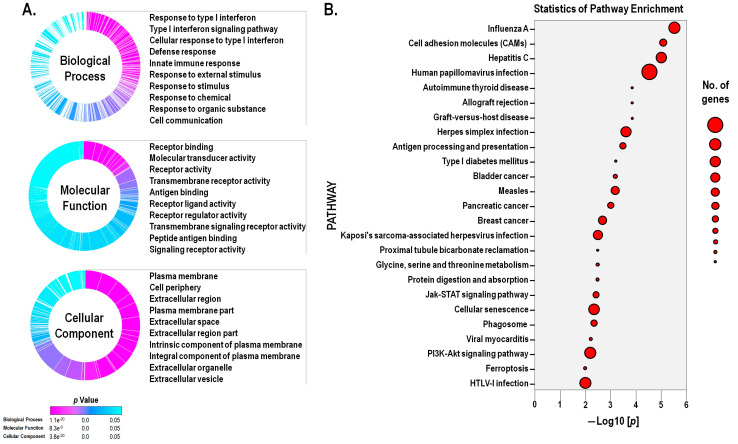
Avicequinone B promoted alterations in the gene expression associated with cellular survival, proliferation, junction and signaling; response to interferon and virus; cancer pathways; and ferroptosis. (A) Differentially expressed genes (DEGs), between avicequinone B (8.20 µM)-treated HT-29 cells and control (vehicle), were categorized within three domains: biological processes, molecular function, and cellular components, through gene ontology (GO) term enrichment analysis performed with iPathwayGuide online software; cut-off p value p <0.05 corrected for multiple comparisons using FDR and Bonferroni. The GO summary graphs plot the significant overrepresented terms for each domain. Each section of the graph represents a different term, the height of the section is proportional to the number of DEGs, and the enrichment significance is displayed using a color scale with cyan being least significant and magenta most significant. (B) Scatter plot of the top twenty-five significantly enriched pathways for DEGs, as categorized by iPathwayGuide online software. Pathways are ranked (x-axis) according to their significance (y-axis) which is represented in terms of the negative log (base 10) of the P value (without correction). The size of the dots represents the number of DEGs.