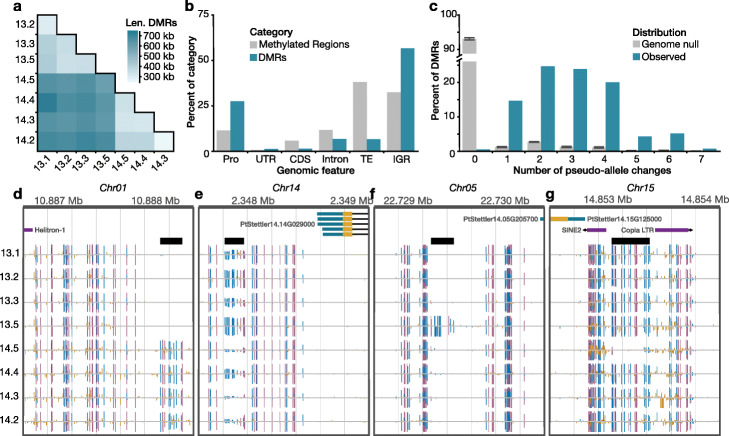
Fig. 4.
Identification and quantification of somatic stability of differentially methylated regions. a Divergence of differentially methylated regions corresponds to divergence in age. The darker color indicates combined length of the pairwise DMRs. b The genome-wide distribution of identified DMRs in genomic features. Abbreviations: TE, transposable elements and repeats; IGR, intergenic region; Pro, promoter region (2 kb upstream of TSS); UTR, untranslated regions; CDS, coding sequence. Methylated regions were identified in as regions methylated in at least one sample. c There are significantly more pseudo-allele changes between the branches at DMRs (blue) compared to the genome-wide null (Wilcoxon rank sum, one-sided, P value < 2 × 10− 16). Gray bars are the genome-wide null as mean +/− std. dev. across 10 simulations. d Browser screenshot of a tree specific DMR where all branches in tree 13 are homozygous unmethylated and all branches of tree 14 are homozygous methylated. e Browser screenshot of a highly variable DMR where the pseudo-allele state changes between each branch. f Browser screenshot of a single gain DMR where all branches except 13.5 are homozygous unmethylated and 13.5 gains methylation. g Browser screenshot of a single loss DMR where all branches except 14.5 are homozygous methylated and 14.5 has lost methylation. For d–g, gene models and transposable elements are shown at the top and methylome tracks are below. Vertical bars indicate methylation at the position, where height corresponds to level and color is context, red for CG, blue for CHG, and yellow for CHH. DMR is indicated by thick black horizontal line