Whole‐genome duplication (WGD) is a dramatic, common event in plants. Because of the detrimental effects arising from doubling the entire chromosome set, such as minority cytotype exclusion, genomic instability, mitotic and meiotic abnormalities, alterations in cell architecture, or epigenetic changes (Comai, 2005), purifying selection is expected to quickly remove polyploids from the population. Nevertheless, polyploidy is common, and many diploids bear traces of a polyploid ancestry (Soltis et al., 2015; Van de Peer et al., 2017). Some of these ancestral polyploidy events can be traced back to the origin of large and diverse taxonomic lineages; also, most crops are neopolyploids, suggesting an important role for WGDs in promoting phenotypic diversity, speciation, and domestication.
Interestingly, the long‐term fixation of polyploidy does not seem to occur randomly. One notable example is the biased distribution of WGD events across independent plant lineages at the Cretaceous–Paleogene or K‐Pg boundary, about 66 million years ago (Ma). Many lineages seem to have undergone a WGD around the time of the K‐Pg extinction event, while their nonpolyploid ancestors died out, suggesting that polyploidy might confer an adaptive advantage under stressful environments and periods of environmental turmoil (Van de Peer et al., 2017). There might be other examples of “waves” of WGDs during periods of environmental change. For instance, the timing of WGD events in the Malpighiales may correlate with periods of global climatic change during the Paleocene–Eocene, ca. 56–54 mya (Cai et al., 2019). However, the evolutionary mechanisms underlying the relationship between WGD and evolutionary success remain elusive, despite vivid discussions on the topic, as witnessed by a recent special series of review papers and several other essays in the “On the Nature of Things” section of this very same journal. Here, we try to reconcile and discuss some recent findings about the putative adaptive role of WGD during evolution under scenarios of global ecological challenge.
WGD AND ITS EVOLUTIONARY IMPACT MAY BE TEMPORALLY UNCOUPLED
Direct evidence linking WGD and “escape from extinction” remains controversial, in part because of the difficulties in unambiguously inferring and dating WGD events (Zwaenepoel et al., 2019). Furthermore, some findings suggest that WGD events can impact evolution long after they occur. For example, across angiosperms, a species‐rich crown group and a species‐poor sister clade often share the same ancestral WGD event. This common pattern suggests that post‐WGD species’ radiations may occur long after the WGD event (Schranz et al., 2012). The model “lineage‐specific ohnologue resolution” (LORe) has recently suggested that delayed rediploidization can explain these “lag times” (Robertson et al., 2017). Delayed rediploidization occurs as long as any two of the four chromosome sets resulting from WGD pair randomly during meiosis, leading to recombination among them and thus preventing sequence and functional divergence between homoeologs to occur (Campbell et al., 2019). Therefore, LORe provides a plausible explanation of how the functional and evolutionary outcomes of genome doubling, and particularly its contribution to extinction avoidance, may be constrained from arising for millions or tens of millions of years after the WGD event (Robertson et al., 2017). An important consequence of the LORe model is also that inferred dates of WGD events might not necessarily coincide with an extinction event in order for the WGD to confer a selective advantage under highly unstable or stressful environments (Fig. 1).
Figure 1.
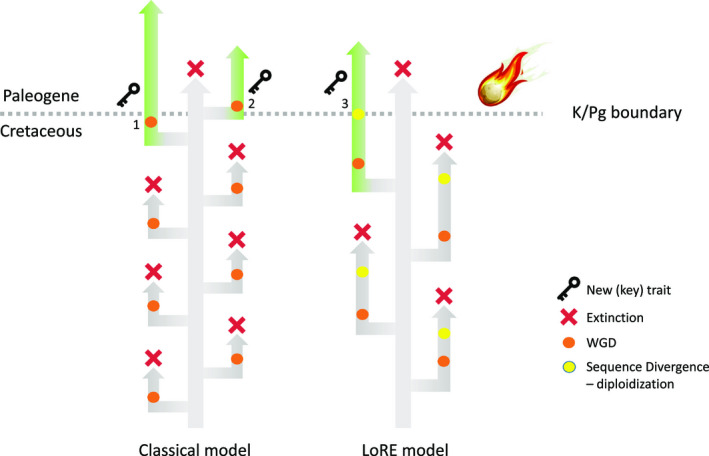
The different outcomes of whole‐genome duplication (WGD). WGDs can get established because of increased genetic variation, in turn increasing the adaptive potential of the species concerned, for instance through the evolution of novel traits (1). WGDs can also occur subsequent to a cataclysmic impact (2), as a result of stress, leading to, for instance, an increased number of unreduced gametes. It has recently been suggested that the WGD events, the extinction event, and the escape from extinction do not need to be coincident in time. For example, the LoRE model (Robertson et al., 2017) assumes that sequences diverge between homoeologs after a WGD event, and thus, the opportunity for evolutionary innovation and adaptation is postponed until rediploidization occurs. As long as there is tetrasomic inheritance, there is recombination and genes, and subgenomes do not diverge. Only after diploidization (leading to disomic inheritance), do both subgenomes start to diverge (as are their genes), and the evolutionary outcome of WGD can be realized, e.g., extinction avoidance (3). Consequently, WGDs with delayed diploidization as predicted by the LoRE model can be dated quite a bit younger than they actually are. Interestingly though, if WGD is promoting adaptation to highly disturbed environments, (e.g., during extinctions), the WGD will also be dated at the time of these events, even if it has occurred earlier. (Figure courtesy of Peter W. H. Holland, University of Oxford, UK).
Whole‐genome duplication, resulting in the duplication of every gene in the genome, is considered a major source of novel raw genetic material upon which mutation and selection may act to evolve novel and/or specialized biological functions with potential adaptive value. However, the potential of WGD in promoting evolutionary change may be constrained by requirements to maintain dosage balance (DB) among gene duplicates connected in biological networks (Conant and Wolfe, 2008; Tasdighian et al., 2017). The loss, or functional divergence, of a connected gene duplicate may upset the stoichiometric balance with other genes connected in the network and is thus expected to be disfavored. Although DB is generally considered as the primary force driving gene‐duplicate retention, DB constraints are expected to relax over time. Again, the observation of significant “lag times” between WGDs and their evolutionary outcome (Schranz et al., 2012) is compatible with a model of DB as a transitional state under which evolution of gene duplicates is constrained immediately after WGD, followed by functional specialization or loss through pseudogenization. These time lags may well overlap with the period subsequent to the WGD event during which divergence of homoeologs is constrained by DB effects. The relaxation of DB constraints over time will also likely be dependent on the ecological requirements of the species (Carretero‐Paulet and Fares, 2012; Conant et al., 2014).
IS INCREASED GENOMIC COMPLEXITY FOLLOWING WGD A HINDRANCE FOR EVOLUTION DURING STABLE TIMES, BUT AN OPPORTUNITY DURING TIMES OF ENVIRONMENTAL TURMOIL?
To unambiguously establish the role of WGD in promoting evolutionary success and adaptation during periods of environmental upheaval, one should be able to “replay the duplication tape of life” in model organisms assessing the effects of WGD on fitness under different selection regimes. Studies in yeast show that polyploidy can accelerate evolutionary adaptation to challenging environments, which could occur through complex effects on the frequency or fitness of beneficial mutations (Selmecki et al., 2015), or because regulatory robustness conferred by WGD allows a wider range of phenotypic responses to environmental stresses (Keane et al., 2014). Unfortunately, attempting similar evolution experiments in plants is difficult and constrained by experimental limitations.
Alternatively, computational approaches aimed at simulating genome evolution under realistic biological models have investigated the potential adaptive role of WGD (see e.g., Cuypers et al., 2017). Recent work suggested that nonpolyploids performed better than polyploids under stable environments, while the opposite holds true for unstable environments (Yao et al., 2019). These observations were interpreted as random mutations causing changes in the genome and its encoded gene regulatory networks (GRNs), an effect that is expected to be exacerbated in more complex GRNs, such as those arising from WGD, featured by a duplicated number of nodes (i.e., genes) and an exponentially increased number of edges (i.e., interactions). Therefore, under stable environments, random mutations in complex GRNs—and thus with effects widely propagated over the network—will often be detrimental or maladaptive (i.e., moving away from peaks in the adaptive/fitness landscape). In contrast, in unstable or stressful environments, these (bigger) changes or disturbances in the GRN may actually allow the drastic adaptive changes necessary to survive (Fig. 2).
Figure 2.
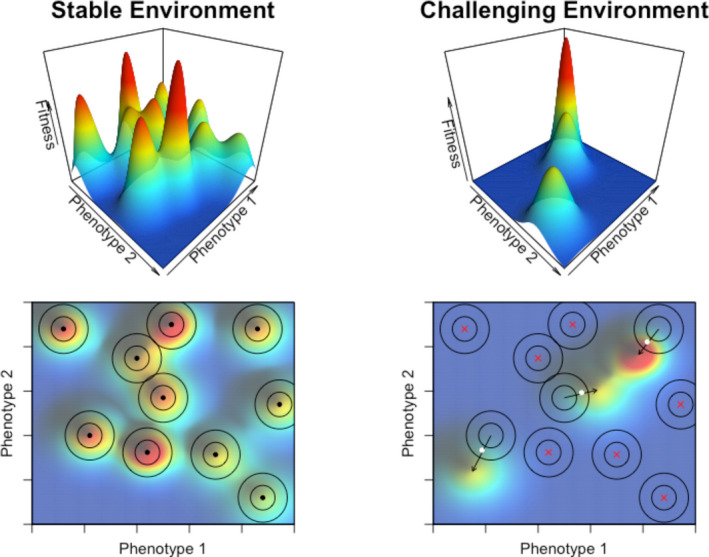
Polyploidy may allow a wider and faster exploration of phenotypic space, ultimately conferring a potential adaptive advantage under challenging environmental conditions. Upper panels, 3D representation of fitness landscapes in which hills, corresponding to local adaptive peaks or niches, are surrounded by valleys or depressions (in dark blue), corresponding to regions of the phenotype space inaccessible to any organism. Bottom panels, contour lines of the fitness landscapes above. Black dots represent organisms that have reached their local adaptive peaks; white dots represent organisms that survived after a shift of their adaptive peaks; red crosses represent organisms that have perished because of big shifts or extinction of their niches. Arrows indicate the trajectories followed by every organism to reach an adaptive peak. Concentric circles around organisms represent the areas of the fitness landscape accessible by their nonpolyploid (inner circle) or polyploid relatives (outer circle). In stable environments (left panels), nonpolyploid organisms are expected to have reached their local adaptive peaks, outcompeting their polyploid relatives, which are more likely to fall farther away. Adaptive landscapes are readily distorted by environmental challenges, such as cataclysmic or extinction events (right panels), resulting in fewer available ecological niches and in big shifts in the relative locations of their adaptive peaks. Under these conditions, although most organisms are expected to perish, polyploid organisms, featured by wider accessible phenotype space (see text for details), have better chances to fall near the peak of a newly formed adaptive hill and thus to acquire the necessary evolutionary innovations to colonize novel niches.
Duplication of entire genomes and their GRNs immediately creates redundant “modules” in the genome. Modules are represented by subnetworks of the global GRN formed by highly connected genes that are co‐expressed and/or co‐regulated by the same set of key regulators, while establishing sparser connections with genes from different modules (Espinosa‐Soto, 2018). The role of gene duplication and cis‐regulatory evolution in rapid GRN rewiring has been reviewed (see e.g., Das Gupta and Tsiantis, 2018). Furthermore, redundant modules created through WGD might facilitate evolutionary innovation and rapid adaption to novel environments, a trait called evolvability, in different ways. To cite a few, first, network redundancy after WGD can help to buffer against deleterious mutations, contributing to the genetic robustness (Keane et al., 2014). Second, by increasing the absolute dosage of all the genes in a pathway, WGD can lead to increased fitness under conditions that require elevated fluxes for certain pathways, as has been illustrated in an elegant simulation experiment in yeast (van Hoek and Hogeweg, 2009). Third, the probabilities of rewiring biological networks formed by multiple connected genes into specialized metabolic, regulatory, or developmental pathways increase if all genes involved duplicate together by means of WGD and co‐evolve partitioned subfunctions, such as interactions partners or expression domains (De Smet and Van de Peer, 2012). Similarly, modularity increases the probabilities of rewiring newly duplicated networks into entirely novel biological pathways, with null or limited effects on the ancestral network, a pattern consistent with network neofunctionalization (Edger et al., 2015). The WGD duplicates, originally retained neutrally through requirements to maintain DB within the concerned subnetwork, can, under certain environmental constraints, contribute to the complex changes at the genomic level and, ultimately, to the phenotypic plasticity required to overcome periods of environmental upheaval. The redundant and modular structure of duplicated GRNs allows both a wider and faster exploration of phenotypic space and resulting fitness landscapes, ultimately offering increased possibilities for evolutionary innovation and adaptation in a (drastically) changed environment.
CONCLUSIONS
The nonrandom distribution of WGD across the “green” tree of life poses an interesting conundrum about their role in evolution. On the one hand, the detrimental effects of WGD, with the requirements to maintain DB among duplicates, are expected to constrain evolutionary innovation or adaptation. On the other hand, many species are currently polyploid or have a polyploid ancestry. Ancient WGDs seem to coincide with disruptive events, such as periods of environmental turmoil or events of extinction (Van de Peer et al., 2017). These observations prompted speculations about a potential adaptive advantage conferred by WGD, strong enough to alleviate its intrinsic disadvantages, at least under extreme environmental constraints.
Here, we reiterated previous observations that increased redundancy and modularity of GRNs encoded by duplicated genomes would allow for a wider—and faster—exploration of phenotypic space. While big leaps in the corresponding adaptive landscape are expected to be selected against under stable environments, under highly disturbed ones they might actually result in successful adaptation and the occupation of a novel adaptive peak (Fig. 2). Assuming a potential advantage of WGD under disturbed environments, it is tempting to speculate on the future of current polyploids. For example, in the context of the ongoing sixth mass extinction with a rate of extinction up to 500 times greater (Humphreys et al., 2019) than the background rate of extinction, should we expect a relative increase in the occurrence of polyploids versus diploids in the major plant lineages (Levin, 2020)? Tackling this question, as well as understanding the molecular mechanisms ultimately responsible for an advantage of duplicated genomes over nonduplicated ones under stressful or disturbed environments, could come from evolutionary competition experiments of polyploid versus nonpolyploid populations of a suitable plant model organism. Appropriate experiments should be designed to reproduce the environmental conditions imposed by past and current massive extinction events, such as the ones occurring at the K‐Pg boundary or, paradoxically, perhaps also these days.
Acknowledgments
Y.V.d.P. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 833522). We thank two anonymous reviewers for their constructive comments and apologize to those whose work could not be cited because of space limitations.
Carretero-Paulet, L. and Van de Peer Y.. 2020. The evolutionary conundrum of whole-genome duplication. American Journal of Botany 107(8): 1101–1105.
Contributor Information
Lorenzo Carretero‐Paulet, Email: lpaulet@ual.es.
Yves Van de Peer, Email: yves.vandepeer@psb.ugent.be.
LITERATURE CITED
- Cai, L. , Xi Z., Amorim A. M., Sugumaran M., Rest J. S., Liu L., and Davis C. C.. 2019. Widespread ancient whole‐genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytologist 221: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, M. A. , Hale M. C., McKinney G. J., Nichols K. M., and Pearse D. E.. 2019. Long‐term conservation of ohnologs through partial tetrasomy following whole‐genome duplication in Salmonidae. G3 9: 2017–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero‐Paulet, L. , and Fares M. A.. 2012. Evolutionary dynamics and functional specialization of plant paralogs formed by whole and small‐scale genome duplications. Molecular Biology and Evolution 29: 3541–3551. [DOI] [PubMed] [Google Scholar]
- Comai, L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews Genetics 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Conant, G. C. , Birchler J. A., and Pires J. C.. 2014. Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Current Opinion in Plant Biology 19: 91–98. [DOI] [PubMed] [Google Scholar]
- Conant, G. C. , and Wolfe K. H.. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Cuypers, T. D. , Rutten J. P., and Hogeweg P.. 2017. Evolution of evolvability and phenotypic plasticity in virtual cells. BMC Evolutionary Biology 17: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta, M. , and Tsiantis M.. 2018. Gene networks and the evolution of plant morphology. Current Opinion in Plant Biology 45: 82–87. [DOI] [PubMed] [Google Scholar]
- De Smet, R. , and Van de Peer Y.. 2012. Redundancy and rewiring of genetic networks following genome‐wide duplication events. Current Opinion in Plant Biology 15: 168–176. [DOI] [PubMed] [Google Scholar]
- Edger, P. P. , Heidel‐Fischer H. M., Bekaert M., Rota J., Glockner G., Platts A. E., Heckel D. G., et al. 2015. The butterfly plant arms‐race escalated by gene and genome duplications. Proceedings of the National Academy of the Sciences USA 112: 8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa‐Soto, C. 2018. On the role of sparseness in the evolution of modularity in gene regulatory networks. PLoS Computational Biology 14: e1006172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, A. M. , Govaerts R., Ficinski S. Z., Lughadha E. N., and Vorontsova M. S.. 2019. Global dataset shows geography and life form predict modern plant extinction and rediscovery. Nature Ecology and Evolution 3: 1043–1047. [DOI] [PubMed] [Google Scholar]
- Keane, O. M. , Toft C., Carretero‐Paulet L., Jones G. W., and Fares M. A.. 2014. Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae. Genome Research 24: 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. A. 2020. Has the polyploid wave ebbed? Frontiers in Plant Science 11: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, F. M. , Gundappa M. K., Grammes F., Hvidsten T. R., Redmond A. K., Lien S., Martin S. A. M., et al. 2017. Lineage‐specific rediploidization is a mechanism to explain time‐lags between genome duplication and evolutionary diversification. Genome Biology 18: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz, M. E. , Mohammadin S., and Edger P. P.. 2012. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag‐Time Model. Current Opinion in Plant Biology 15: 147–153. [DOI] [PubMed] [Google Scholar]
- Selmecki, A. M. , Maruvka Y. E., Richmond P. A., Guillet M., Shoresh N., Sorenson A. L., De S., et al. 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis, P. S. , Marchant D. B., Van de Peer Y., and Soltis D. E.. 2015. Polyploidy and genome evolution in plants. Current Opinion in Genetics and Development 35: 119–125. [DOI] [PubMed] [Google Scholar]
- Tasdighian, S. , Van Bel M., Li Z., Van de Peer Y., Carretero‐Paulet L., and Maere S.. 2017. Reciprocally retained genes in the angiosperm lineage show the hallmarks of dosage balance sensitivity. Plant Cell 29: 2766–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y. , Mizrachi E., and Marchal K.. 2017. The evolutionary significance of polyploidy. Nature Reviews Genetics 18: 411–424. [DOI] [PubMed] [Google Scholar]
- van Hoek, M. J. , and Hogeweg P.. 2009. Metabolic adaptation after whole genome duplication. Molecular Biology and Evolution 26: 2441–2453. [DOI] [PubMed] [Google Scholar]
- Yao, Y. , Carretero‐Paulet L., and Van de Peer Y.. 2019. Using digital organisms to study the evolutionary consequences of whole genome duplication and polyploidy. PLoS One 14: e0220257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel, A. , Li Z., Lohaus R., and Van de Peer Y.. 2019. Finding evidence for whole genome duplications: a reappraisal. Molecular Plant 12: 133–136. [DOI] [PubMed] [Google Scholar]


