Abstract
This study aims to uncover the potential function of MicroRNA-543 (miRNA-543) in the pathogenesis of gastric carcinoma and the possible mechanism. MiRNA-543 levels in gastric carcinoma tissues and cell lines were detected by quantitative real-time polymerase chain reaction (qRT-PCR). Regulatory effects of miRNA-543 on proliferative and migratory abilities of AGS and MKN45 cells were assessed. The downstream target of miRNA-543 was predicted by online bioinformatics and verified by dual-luciferase reporter gene assay. At last, rescue experiments were carried out to uncover the interaction between miRNA-543 and Krüppel-like factor 6 (KLF6) in the progression of gastric carcinoma. MiRNA-543 was upregulated in gastric carcinoma tissues and cell lines. Particularly, gastric carcinoma patients with advanced stage or positive metastasis expressed higher abundance of miRNA-543. Overexpression of miRNA-543 promoted proliferative ability in gastric carcinoma, manifesting as increased viability, EdU-positive ratio and migratory cell number in AGS and MKN45 cells. KLF6 was proved to be the downstream target of miRNA-543. Both mRNA and protein levels of KLF6 were negatively regulated by miRNA-543 in gastric carcinoma cells. Silence of KLF6 was able to reverse the regulatory effects of miRNA-543 inhibitor on proliferative and migratory abilities in gastric carcinoma. MiRNA-543 is highly expressed in gastric carcinoma. It accelerates gastric carcinoma cells to proliferate and migrate by negatively regulating KLF6 level.
Keywords: Gastric carcinoma, miRNA-543, KLF6, proliferation, migration
Introduction
Gastric carcinoma is a common malignancy derived from the gastric mucosal epithelium [1]. Studies have confirmed that the occurrence of gastric carcinoma is the result of both host genetic factors and environmental risk factors. Risk factors for tumorigenesis of gastric include demographic characteristics, dietary patterns, microbial infections, genetic differences, and mental and psychological factors [2]. Surgical resection is currently considered to be the only possible curative treatment for gastric carcinoma. Despite great advances in cancer screening and treatment technologies, the 5-year survival of gastric carcinoma is lower than 30%. It is noteworthy that the 5-year survival in patients with early-stage gastric carcinoma undergoing radical resection is up to 90%. Nevertheless, the detective rate of early-stage gastric carcinoma is extremely low owing to the lack of typical symptoms, extensive application of gastroscopy and public medical knowledge [3,4]. Advanced gastric carcinoma is fatal and its prognosis is extremely poor. It is urgent to develop biological markers and therapeutic targets for gastric carcinoma [5].
MicroRNAs (miRNAs) are endogenous, non-coding RNAs of 21-25 nucleotides in length. A pri-miRNA is transcribed in the nucleus, which in turn produces a pre-miRNA with 60-70 nucleotides long and contains a hairpin structure. Subsequently, a pre-miRNA is translocated into the cytoplasm under the assistance of Ran-GTP, where a specific RNA-specific endonuclease Dicer contributes to process a double-stranded RNA. This short-chain double-stranded RNA is separated by the RNA helicase, one of which is integrated into RISC (RNA-induced silencing complex) and the other one is discarded. At last, the RISC-miRNA complex splices or inhibits translation of target RNAs by complementary base pairing, thus downregulating downstream protein expressions [6,7]. Therefore, the balance between mRNA and miRNA controls mRNA abundance to ensure normal expression pattern of proteins. Once the balance is disrupted, miRNAs exert oncogenic or anti-tumor effects in tumor diseases [8-12].
MiRNA-543 is highly expressed in various malignant tumors [13-16]. Overexpression of miRNA-543 in renal clear cell carcinoma promotes tumor cells to proliferate and migrate, presenting a carcinogenic role [13]. MiRNA-543 regulates multiple tumor-associated genes, including Krüppel-like factor 6 (KLF6) and Silent information regulator T1 (SIRT1) [17,18]. This experiment mainly elucidated the role of miRNA-543 in the development of gastric carcinoma.
Patients and methods
Sample collection
Fifty gastric carcinoma tissues and matched adjacent tissues were collected. None of gastric carcinoma patients enrolled was treated with preoperative anti-tumor therapy. Samples were pathologically confirmed and preserved in liquid nitrogen. Patients and their families in this study have been fully informed. This study was approved by Ethics Committee of the First Medical Centre, Chinese PLA General Hospital.
Cell culture
Gastric carcinoma cells (MKN45, AGS, SNU5 and HGC-27) and gastric mucosal cells (GES1) were cultured in dulbecco’s modified eagle medium (DMEM) (Gibco, Rockville, MD, USA) containing 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA), and 1% penicillin-streptomycin in an incubator containing 5% CO2 at 37°C. Fresh medium was regularly replaced.
Transfection
Cells in good growth condition were transfected using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Transfected cells for 24-48 h were harvested for the following experiments. Transfection plasmids used were constructed by Genechem (Shanghai, China).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Cells were lysed to harvest RNAs using TRIzol method (Invitrogen, Carlsbad, CA, USA), and the extracted RNAs were subjected to reverse transcription according to the instructions of PrimeScript RT reagent Kit (TaKaRa, Tokyo, Japan). RNA concentration was detected using a spectrometer. QRT-PCR was then performed based on the instructions of SYBR Premix Ex Taq TM (TaKaRa, Tokyo, Japan). Relative level was calculated using 2-ΔΔCt method. miRNA-543, F: 5’-CCAGCTACACTGGGCAGCAGCAATTCATGTTT-3’; R: 5’-CTCAACTGGTGTCGTGGA-3’; KLF6, F: 5’-CTCTCAGCCTGGAAGCTTTTAGCCTAC-3’; R: 5’-ACAGCTCCGAGGAACTTTCTCCCA-3’; glyceraldehyde 3-phosphate dehydrogenase (GAPDH), F: 5’-GGAATCCACTGGCGTCTTCA-3’; R: 5’-GGTTCACGCCCATCACAAAC-3’ U6, F: 5’-AGAGAAGATTAGCATGGCCCCTG-3’; R: 5’-ATCCAGTGCGGGTCCGAGG-3’.
5-Ethynyl-2’-deoxyuridine (EdU) assay
Cells were inoculated into 96-well plates with 1×105 cells per well, and labeled with 100 μL of EdU reagent (50 μM) per well for 2 h. After washing with phosphate buffered saline (PBS), cells were fixed in 50 μL of fixation buffer, decolored with 2 mg/mL glycine and permeated with 100 μL of penetrant. After washing with PBS once, cells were stained with 4’,6-diamidino-2-phenylindole (DAPI) in dark for 30 min. EdU-positive ratio was determined under a fluorescent microscope.
Cell counting kit-8 (CCK-8) assay
Cells were seeded into 96-well plates with 5×104 cells per well. At the appointed time points, 10 μL of CCK-8 solution (Dojindo, Kumamoto, Japan) was added in each well. The absorbance at 450 nm of each sample was measured by a microplate reader (Bio-Rad, Hercules, CA, USA).
Transwell
1×105 cells were inoculated in the upper side of 8 μm Transwell chamber (Corning, Corning, NY, USA) coated with diluted Matrigel. In the bottom side, 600 μL of medium containing 20% FBS was applied. After 48 h of incubation, cells penetrated to the bottom side were fixed in 4% paraformaldehyde for 20 min, stained with crystal violet for 20 min and counted using a microscope. The number of penetrating cells was counted in 5 randomly selected fields per sample.
Dual-luciferase reporter gene assay
Based on the predicted binding sites between miRNA-543 and KLF6, luciferase vectors KLF6-WT and KLF6-MUT were constructed. Cells were co-transfected with NC/miRNA-543 mimics and KLF6-WT/KLF6-MUT. Finally, cells were lysed for examining relative luciferase activity (Promega, Madison, WI, USA).
Western blot
Total protein was extracted for determining protein concentration. Protein sample was separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride (PVDF) membranes (Roche, Basel, Switzerland) and blocked in 5% skim milk for 1 hour. The specific primary antibody was used to incubate with the membrane overnight at 4°C, followed by secondary antibody incubation for 2 h at room temperature. After washing with 1×tris buffered saline-tween (TBST) for 1 min, the chemiluminescent substrate kit was used for exposure of the protein bands.
Statistical analysis
Statistical Product and Service Solutions (SPSS) 22.0 (IBM, Armonk, NY, USA) and Graphpad prism 8.0 (La Jolla, CA, USA) was used for data analysis and figure editing. Data were expressed as mean ± standard deviation (x̅ ± SD). Pearson correlation test was conducted to compare the relationship between two genes. Intergroup data were compared using the t-test. P<0.05 considered the difference was statistically significant.
Results
Upregulation of miRNA-543 in gastric carcinoma
MiRNA-543 levels in gastric carcinoma tissues and adjacent normal tissues were detected at first, which was upregulated in tumor tissues compared with normal tissues (Figure 1A). According to the tumor staging, we determined miRNA-543 level in gastric carcinoma patients in stage I+II and stage III+IV. Patients in stage III+IV expressed higher abundance of miRNA-543 compared with stage I+II (Figure 1B). In addition, gastric carcinoma patients with positive metastasis expressed higher level of miRNA-543 relative to those without metastases (Figure 1C). In vitro level of miRNA-543 was identically upregulated in gastric carcinoma cells than that of gastric mucosal cells (Figure 1D). The above data indicated the potential involvement of miRNA-543 in the progression of gastric carcinoma.
Figure 1.
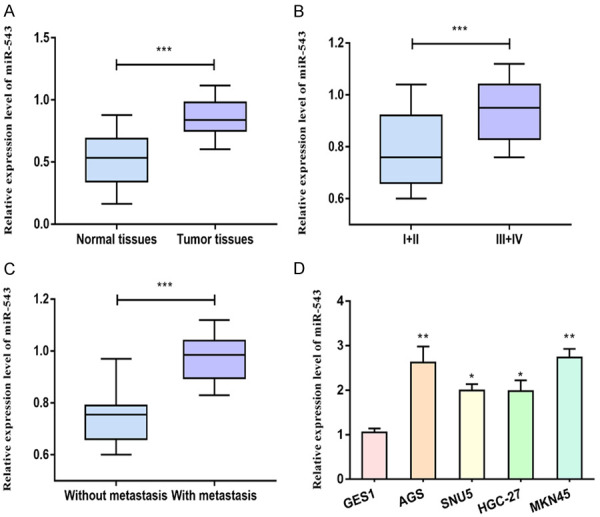
Upregulation of miRNA-543 in gastric carcinoma. A. MiRNA-543 level in gastric carcinoma tissues and adjacent normal tissues. B. MiRNA-543 level in gastric carcinoma patients with stage I+II and stage III+IV. C. MiRNA-543 level in gastric carcinoma patients with negative or positive metastasis. D. MiRNA-543 level in gastric carcinoma cells and gastric mucosal cells.
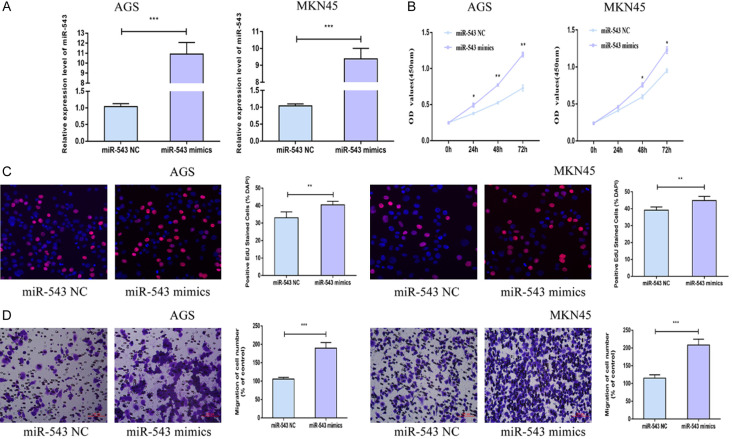
MiRNA-543 stimulated gastric carcinoma to proliferate and migrate
In vitro overexpression model of miRNA-543 was established in AGS and MKN45 cells by transfection of miRNA-543 mimics (Figure 2A). CCK-8 assay uncovered an elevated viability in gastric carcinoma cells overexpressing miRNA-543 compared with miRNA-543 NC group (Figure 2B). Meanwhile, EdU-positive ratio increased after transfection of miRNA-543 mimics, also confirming the stimulated proliferative ability compared with miRNA-543 NC group (Figure 2C). Transwell assay illustrated an increase in migratory cell number after overexpression of miRNA-543 in AGS and MKN45 cells in comparison with miRNA-543 NC group (Figure 2D). Hence, miRNA-543 was able to promote gastric carcinoma to proliferate and migrate.
Figure 2.
MiRNA-543 stimulated gastric carcinoma to proliferate and migrate. A. Transfection efficacy of miRNA-543 mimics in AGS and MKN45 cells. B. Viability in AGS and MKN45 cells transfected with NC or miRNA-543 mimics. C. EdU-positive ratio in AGS and MKN45 cells transfected with NC or miRNA-543 mimics (magnification 20×). D. Migratory cell number in AGS and MKN45 cells transfected with NC or miRNA-543 mimics (magnification 20×).
MiRNA-543 directly targeted on KLF6
Through online prediction and analyses, KLF6 was selected as the downstream gene of miRNA-543 (Figure 3A). A decline in luciferase activity after co-transfection of KLF6-WT and miRNA-543 mimics verified the binding relationship between KLF6 and miRNA-543 (Figure 3B). Both mRNA and protein levels of KLF6 were downregulated in AGS and MKN45 cells overexpressing miRNA-543 compared with miRNA-543 NC group (Figure 3C, 3D). Furthermore, KLF6 level was identified to be downregulated in gastric carcinoma tissues compared with normal tissues (Figure 3E). Through statistical processing, miRNA-543 level was negatively correlated to that of KLF6 in gastric carcinoma tissues (Figure 3F). It is proved that KLF6 was the downstream gene of miRNA-543 and negatively regulated by it in gastric carcinoma.
Figure 3.
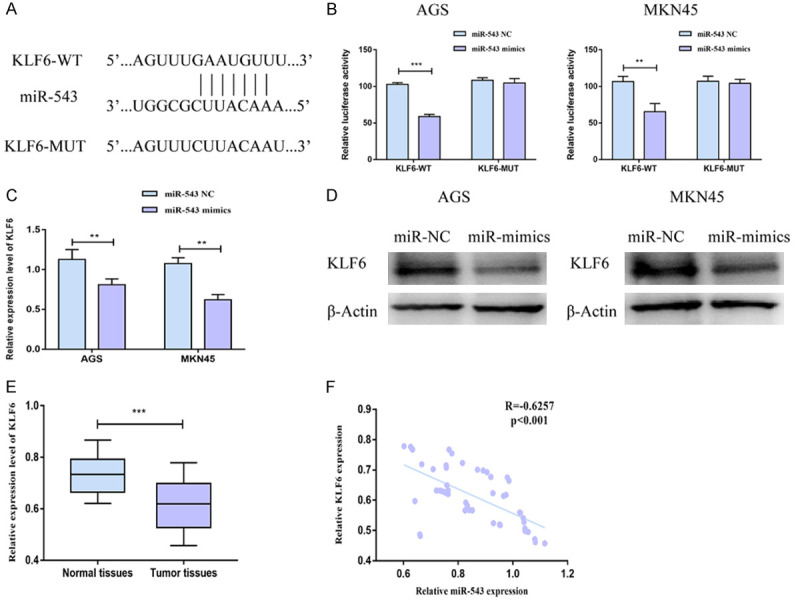
MiRNA-543 directly targeted on KLF6. (A) Binding sites in promoter regions of miRNA-543 and KLF6. (B) Luciferase activity in AGS and MKN45 cells co-transfected with NC/miRNA-543 mimics and KLF6-WT/KLF6-MUT. (C, D) The mRNA (C) and protein (D) levels of KLF6 in AGS and MKN45 cells transfected with NC or miRNA-543 mimics. (E) KLF6 level in gastric carcinoma tissues and adjacent normal tissues. (F) A negative correlation between expression levels of miRNA-543 and KLF6 in gastric carcinoma tissues.
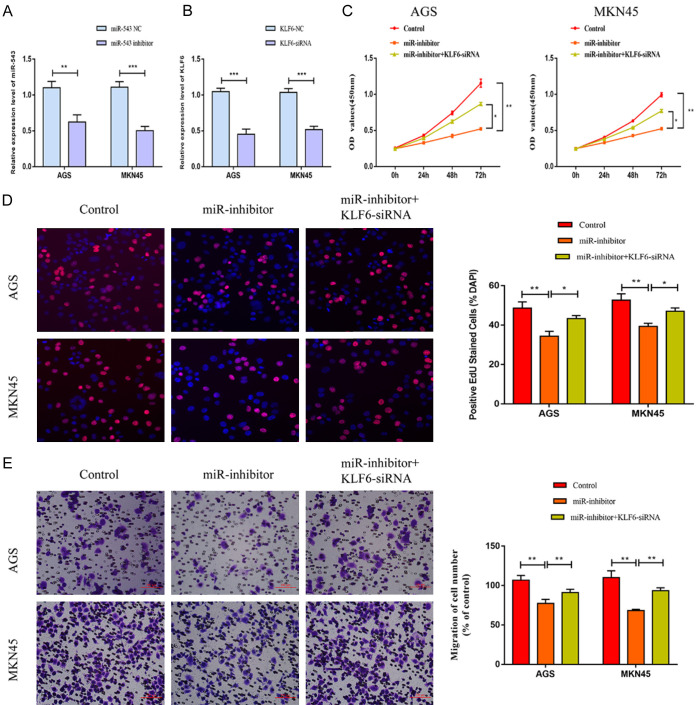
KLF6 was responsible for miRNA-543 regulation on gastric carcinoma
To uncover the involvement of KLF6 in the progression of gastric carcinoma, KLF6-siRNA was established. MiRNA-543 inhibitor significantly down-regulated the level of miRNA-543 in AGS and MKN45 cells. Transfection of KLF6-siRNA remarkably down-regulated KLF6 level in AGS and MKN45 cells (Figure 4A, 4B). Knockdown of miRNA-543 reduced viability and EdU-positive ratio in gastric carcinoma, which were partially reversed after co-transfection of KLF6-siRNA (Figure 4C, 4D). Besides, the attenuated migration in gastric carcinoma cells with miRNA-543 knockdown was enhanced after silence of KLF6 (Figure 4E). It is suggested that KLF6 was necessary in the gastric carcinoma progression regulated by miRNA-543.
Figure 4.
KLF6 was responsible for miRNA-543 regulation on gastric carcinoma. AGS and MKN45 cells were transfected with control, miRNA-543 inhibitor or miRNA-543 inhibitor + KLF6-siRNA. A. MiRNA-543 level; B. KLF6 level; C. Viability; D. EdU-positive ratio (magnification 20×); E. Migratory cell number (magnification 20×).
Discussion
MiRNAs are of clinical value in gastric carcinoma, serving as detective indicators, therapeutic targets or prognostic hallmarks. Compared with other circulating nucleic acids, peripheral blood levels of miRNAs are more stable [19,20]. Drug therapy based on tumor-related miRNAs are promising in nowadays tumor treatment. On the one hand, these miRNAs contribute to suppress tumor-related genes and pathways, thus alleviating the malignant progression [21]. On the other hand, miRNAs are able to influence drug-resistance in chemotherapy or radiotherapy through targeting drug transporters, drug-metabolizing enzymes, transcription factors, and nuclear receptors [22]. Dysfunctional miRNAs in gastric carcinoma pose a huge impact on cellular behaviors of tumor cells [23]. Expression characteristics of these miRNAs are closely linked to tumor staging, malignant progression and clinical outcome of gastric carcinoma [24].
KLF6 is a zinc finger transcription factor of the Kruppel-like factor family. It is involved in the regulation of various life activities. In NSCLC and renal clear cell carcinoma, KLF6 is found to be downregulated and induces apoptosis [25,26]. Through p53-independent pathway, KLF6 regulates p21 level and further stimulates tumor cell apoptosis. Once KLF6 is abnormally expressed in tumors, p21 dysfunction leads to apoptosis impair. The malignant proliferative stage of tumor cells eventually leads to tumor deterioration [27,28].
In this paper, miRNA-543 was highly expressed in gastric carcinoma tissues and cell lines. Its level was observed to be higher in gastric carcinoma patients with advanced stage or positive metastasis. In vitro experiments confirmed the role of miRNA-543 to promote proliferative ability in AGS and MKN45 cells. Subsequently, KLF6 was proved to be the downstream target of miRNA-543. Both mRNA and protein levels of KLF6 were negatively regulated by miRNA-543 in gastric carcinoma cells. Silence of KLF6 was able to reverse the regulatory effects of miRNA-543 inhibitor on proliferative and migratory abilities in gastric carcinoma. To sum up, a regulatory loop miRNA-543/KLF6 was identified, which influenced the malignant progression of gastric carcinoma.
Conclusions
MiRNA-543 is highly expressed in gastric carcinoma. It accelerates gastric carcinoma cells to proliferate and migrate by negatively regulating KLF6 level.
Disclosure of conflict of interest
None.
References
- 1.Liou JM, Lee YC, El-Omar EM, Wu MS. Efficacy and long-term safety of H. pylori eradication for gastric cancer prevention. Cancers (Basel) 2019;11:593. doi: 10.3390/cancers11050593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26–38. doi: 10.5114/pg.2018.80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coutzac C, Pernot S, Chaput N, Zaanan A. Immunotherapy in advanced gastric cancer, is it the future? Crit Rev Oncol Hematol. 2019;133:25–32. doi: 10.1016/j.critrevonc.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Hosoda K, Watanabe M, Yamashita K. Re-emerging role of macroscopic appearance in treatment strategy for gastric cancer. Ann Gastroenterol Surg. 2019;3:122–129. doi: 10.1002/ags3.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, Bleotu C, Diaconu CC, Chivu-Economescu M. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029–2044. doi: 10.3748/wjg.v25.i17.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musavi SM, Eghbal-Fard S, Mehrisofiani V, Abd YN, Rahbar FO, Marofi F, Yousefi M. MicroRNAs and signaling networks involved in epithelial-mesenchymal transition. J Cell Physiol. 2019;234:5775–5785. doi: 10.1002/jcp.27489. [DOI] [PubMed] [Google Scholar]
- 8.Wang F. miR-384 targets metadherin gene to suppress growth, migration, and invasion of gastric cancer cells. J Int Med Res. 2019;47:926–935. doi: 10.1177/0300060518817171. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Liu F, Di Wang X. miR-150-5p represses TP53 tumor suppressor gene to promote proliferation of colon adenocarcinoma. Sci Rep. 2019;9:6740. doi: 10.1038/s41598-019-43231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zan Y, Wang B, Liang L, Deng Y, Tian T, Dai Z, Dong L. MicroRNA-139 inhibits hepatocellular carcinoma cell growth through down-regulating karyopherin alpha 2. J Exp Clin Cancer Res. 2019;38:182. doi: 10.1186/s13046-019-1175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Cui Y, Wang D, Wang Y, Wang L. MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;509:647–656. doi: 10.1016/j.bbrc.2018.12.089. [DOI] [PubMed] [Google Scholar]
- 12.Huang X, Tang F, Weng Z, Zhou M, Zhang Q. MiR-591 functions as tumor suppressor in breast cancer by targeting TCF4 and inhibits Hippo-YAP/TAZ signaling pathway. Cancer Cell Int. 2019;19:108. doi: 10.1186/s12935-019-0818-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ZY, Du Y, Wang L, Liu XH, Guo J, Weng XD. MiR-543 promotes cell proliferation and metastasis of renal cell carcinoma by targeting Dickkopf 1 through the Wnt/beta-catenin signaling pathway. J Cancer. 2018;9:3660–3668. doi: 10.7150/jca.27124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao H, Diao C, Wang X, Xie Y, Liu Y, Gao X, Han J, Li S. MiR-543 promotes migration, invasion and epithelial-mesenchymal transition of esophageal cancer cells by targeting phospholipase A2 group IVA. Cell Physiol Biochem. 2018;48:1595–1604. doi: 10.1159/000492281. [DOI] [PubMed] [Google Scholar]
- 15.Du Y, Liu XH, Zhu HC, Wang L, Ning JZ, Xiao CC. MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell Physiol Biochem. 2017;41:1135–1146. doi: 10.1159/000464120. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Guo X, Feng X, Wang T, Hu Z, Que X, Tian Q, Zhu T, Guo G, Huang W, Li X. MiRNA-543 promotes osteosarcoma cell proliferation and glycolysis by partially suppressing PRMT9 and stabilizing HIF-1alpha protein. Oncotarget. 2017;8:2342–2355. doi: 10.18632/oncotarget.13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke N, Pi LH, Liu Q, Chen L. Long noncoding RNA SNHG7 inhibits high glucose-induced human retinal endothelial cells angiogenesis by regulating miR-543/SIRT1 axis. Biochem Biophys Res Commun. 2019;514:503–509. doi: 10.1016/j.bbrc.2019.04.141. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Ma J, Tang Q, Zhang W, Fu Q, Sun J, Wang H, Song B. MicroRNA-543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Kruppel-like factor 6. Biomed Pharmacother. 2018;97:616–623. doi: 10.1016/j.biopha.2017.10.136. [DOI] [PubMed] [Google Scholar]
- 19.Bai SY, Ji R, Wei H, Guo QH, Yuan H, Chen ZF, Wang YP, Liu Z, Yang XY, Zhou YN. Serum miR-551b-3p is a potential diagnostic biomarker for gastric cancer. Turk J Gastroenterol. 2019;30:415–419. doi: 10.5152/tjg.2019.17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X, Li X, Zhao H, Zhou D, Sun B, Liu A, Zhang J, Cui Z, Ma X, Yuan L. Serum miR-515-3p, a potential new RNA biomarker, is involved in gastric carcinoma. J Cell Biochem. 2019;120:15834–15843. doi: 10.1002/jcb.28854. [DOI] [PubMed] [Google Scholar]
- 21.Guo H, Ji F, Zhao X, Yang X, He J, Huang L, Zhang Y. MicroRNA-371a-3p promotes progression of gastric cancer by targeting TOB1. Cancer Lett. 2019;443:179–188. doi: 10.1016/j.canlet.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Ye T, Yang M, Huang D, Wang X, Xue B, Tian N, Xu X, Bao L, Hu H, Lv T, Huang Y. MicroRNA-7 as a potential therapeutic target for aberrant NF-kappaB-driven distant metastasis of gastric cancer. J Exp Clin Cancer Res. 2019;38:55. doi: 10.1186/s13046-019-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parvaee P, Sarmadian H, Khansarinejad B, Amini M, Mondanizadeh M. Plasma level of microRNAs, miR-107, miR-194 and miR-210 as potential biomarkers for diagnosis intestinal-type gastric cancer in human. Asian Pac J Cancer Prev. 2019;20:1421–1426. doi: 10.31557/APJCP.2019.20.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui HW, Han WY, Hou LN, Yang L, Li X, Su XL. miR-1915-3p inhibits Bcl-2 expression in the development of gastric cancer. Biosci Rep. 2019;39:BSR20182321. doi: 10.1042/BSR20182321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F, Zhang Q, Gong Y, Yu J. The lncKLF6/KLF6 feedback loop regulates the growth of non-small cell lung cancer. Am J Cancer Res. 2018;8:1427–1439. [PMC free article] [PubMed] [Google Scholar]
- 26.Lei Z, Ma X, Li H, Zhang Y, Gao Y, Fan Y, Li X, Chen L, Xie Y, Chen J, Wu S, Tang L, Zhang X. Up-regulation of miR-181a in clear cell renal cell carcinoma is associated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. Urol Oncol. 2018;36:23–93. doi: 10.1016/j.urolonc.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 27.Lang UE, Kocabayoglu P, Cheng GZ, Ghiassi-Nejad Z, Munoz U, Vetter D, Eckstein DA, Hannivoort RA, Walsh MJ, Friedman SL. GSK3beta phosphorylation of the KLF6 tumor suppressor promotes its transactivation of p21. Oncogene. 2013;32:4557–4564. doi: 10.1038/onc.2012.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narla G, Kremer-Tal S, Matsumoto N, Zhao X, Yao S, Kelley K, Tarocchi M, Friedman SL. In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene. 2007;26:4428–4434. doi: 10.1038/sj.onc.1210223. [DOI] [PubMed] [Google Scholar]