Summary
Creating true‐breeding lines is a critical step in plant breeding. Novel, completely homozygous true‐breeding lines can be generated by doubled haploid technology in single generation. Haploid induction through modification of the centromere‐specific histone 3 variant (CENH3), including chimeric proteins, expression of non‐native CENH3 and single amino acid substitutions, has been shown to induce, on outcrossing to wild type, haploid progeny possessing only the genome of the wild‐type parent, in Arabidopsis thaliana. Here, we report the characterization of 31 additional EMS‐inducible amino acid substitutions in CENH3 for their ability to complement a knockout in the endogenous CENH3 gene and induce haploid progeny when pollinated by the wild type. We also tested the effect of double amino acid changes, which might be generated through a second round of EMS mutagenesis. Finally, we report on the effects of CRISPR/Cas9‐mediated in‐frame deletions in the αN helix of the CENH3 histone fold domain. Remarkably, we found that complete deletion of the αN helix, which is conserved throughout angiosperms, results in plants which exhibit normal growth and fertility while acting as excellent haploid inducers when pollinated by wild‐type pollen. Both of these technologies, CRISPR mutagenesis and EMS mutagenesis, represent non‐transgenic approaches to the generation of haploid inducers.
Keywords: CENH3, CENP‐A, haploids, CRISPR, Cas9, αN helix
Introduction
Stable propagation of crop varieties through seeds requires the production of lines homozygous for all relevant traits. In a classical approach, these true‐breeding lines are obtained by self‐pollination or backcrossing for 7–8 generations. With doubled haploid technology, complete homozygosity can be obtained in a single generation (Dwivedi et al., 2015). Haploids are obtained either by in vitro technology or by in vivo technology (Dunwell, 2010; Ren et al., 2017). In vitro haploid induction methods such as anther culture/microspore embryogenesis are expensive and labour‐intensive. Also, many species are recalcitrant to anther culture (Segui‐Simarro et al., 2011). In vivo technologies such as interspecific crosses, severe irradiation of pollen and parthenogenesis are limited to specific crop species. Seed‐based in vivo haploid induction approaches are less labour‐intensive and relatively inexpensive (although many still require in vitro embryo rescue). These include interspecific crosses [i.e. between Hordeum vulgare and Hordeum bulbosum (Ishii et al., 2016; Liu et al., 2016), maize stock 6 and, in potato, the Solanum tuberosum group Phureja haploid inducer clones (Touraev et al., 2009)]. Recently, the genes responsible of haploid induction in maize stock 6 have been identified as a defect in Matrilineal, a sperm‐specific phospholipase (Gilles et al., 2017; Kelliher et al., 2017; Liu et al., 2017). Editing of the orthologous locus in rice has led to haploid induction (Yao et al., 2018), indicating that this approach is not limited to maize. More recently, it was found that mutation of a non‐stock 6‐originating gene ZmDMP enhances haploid induction rate in maize lines that carry stock 6 mutation (Zhong et al., 2019). However, matrilineal is a monocot‐specific phospholipase not found in most dicot species. Recently, it has been shown that egg‐cell‐specific expression of BABY BOOM gene leads to the development of haploid embryos from unfertilized eggs (Khanday et al., 2019). However, this particular approach requires the creation of a new transgenic parent for each segregating line of interest.
With the last decade, the centromeric histone CENH3 has become a target of interest for haploid induction. The basic structure of nucleosome is an octamer of histone proteins composed of two subunits each of histones H2a, H2b, H3 and H4, around which the DNA wraps (Fudenberg and Mirny, 2012). All eukaryotes express multiple H3 variants (Luger, 2003); in Arabidopsis thaliana, there are fifteen (Cui et al., 2015). In the centromeric region, more generic H3 nucleosomes are interspersed with nucleosomes carrying a centromere‐specific H3 variant, termed CENH3 in plants (CENP‐A in humans, CID in flies and CSE4 in yeast) (Earnshaw and Rothfield, 1985; Henikoff et al., 2000; Stoler et al., 1995; Talbert et al., 2002b). CENH3 epigenetically determines the location of centromeres (Allshire and Karpen, 2008), and the simple presence of CENH3 is sufficient to attract additional inner kinetochore proteins. The assembly of inner and outer kinetochore proteins is critical in building a functional kinetochore which in turn affects the binding of spindle fibres and proper segregation of chromosomes (Fukagawa and Earnshaw, 2014). Because of this essential role in cell division, CENH3 (and its other eukaryotic homologs) is an essential protein. Although plant homologs of the majority of kinetochore proteins (known from other species) are hard to identify (Lermontova et al., 2014), and few mutants have been characterized, knockout of the A. thaliana homolog of the centromere‐localized KNL2 protein, involved in CENH3 deposition at centromeres, has been shown to cause a variety of mitotic and meiotic defects (Lermontova et al., 2013), though not lethality.
Ravi and Chan first demonstrated that haploid‐inducing lines (producing tissue culture‐free, seed‐based haploids derived from the wild‐type parent) can be obtained in A. thaliana by substantially modifying CENH3 (Ravi and Chan, 2010; Ravi et al., 2010). Their HI‐inducing modification involved adding an N‐terminal GFP tag to a fusion of the histone 3.3 tail with the CENH3 histone fold domain. This chimeric allele was termed GFP–tailswap. Plants carrying this transgene, in the absence of a wild‐type allele, were male sterile; attempts to rescue this line through outcrossing with wild‐type pollen revealed that the plants produced haploids and aneuploids among their progeny when outcrossed.
Since Ravi and Chan’s discovery of CENH3‐based haploid induction, several modifications in CENH3 have been shown to induce haploids in A. thaliana. Complementation of an A. thaliana CENH3 null mutation with transgenic CENH3 from other species, including Lepidium oleraceum and Brassica rapa, has been shown to induce haploids upon crossing by wild‐type CENH3 pollen (Maheshwari et al., 2015). Six amino acid substitutions in the conserved amino acid residues of CENH3 histone fold domain have been shown to produce normal, fully fertile plants that act as haploid inducers on outcrossing (Karimi‐Ashtiyani et al., 2015; Kuppu et al., 2015).
We set out to identify additional amino acid substitutions that are able to complement, as transgenes, an endogenous cenh3 null but are also abnormal enough to affect chromosome segregation during the first few divisions of early embryogenesis upon outcrossing, leading to genome elimination and haploid induction at a higher rate. Our goal was primarily to identify a wider variety of mutations that might make useful HI inducers if translated to crop plants. However, the generation of these mutants also allows us to distinguish between regions and/or amino acids that are conserved because they are required for CENH3 function in mitosis and meiosis, and those that are conserved because they are required for embryonic and/or endosperm survival on outcrossing, haploid induction being one expression of what has come to be termed a ‘competitive’ relationship between the two sets of centromeres that form the zygote. In an earlier publication, we found that 7 out of 7 amino acid substitutions at highly conserved sites in the histone fold domain (HFD) of CENH3 (Kuppu et al., 2015) resulted in plants with normal phenotypes in terms of their growth and fertility on self‐pollination, but 5 of these experienced a significant ‘outcrossing penalty’ when fertilized with CENH3 wild‐type pollen, and produced haploid progeny among the surviving seeds. This suggests that the outcrossing penalty (dysfunction only on outcrossing to wild type) rather than general mitotic or meiotic centromeric dysfunction is responsible for the highly conserved nature of these particular amino acids.
In this paper, we analyse an additional 31 EMS‐inducible point mutations, all at residues that are highly conserved (within angiosperms), for their ability to perform the mitotic and meiotic activities of CENH3 and their proclivity to induce haploids and/or embryonic death when pollinated by wild‐type pollen. We also study the effect of combining two such point mutations. Finally, in a surprising discovery, we find that CRISPR/Cas9‐mediated in‐frame deletions in the highly conserved αN helix of the CENH3 HFD—including loss of the entire helix—can complement an endogenous null cenh3, producing fully fertile plants that act as a powerful haploid inducers on outcrossing. These findings are valuable to crop breeders as they provide additional options for creating haploid inducers in crop plants. They also provide insights into the remarkable and paradoxical structural flexibility of this extremely conserved protein domain.
Results
The majority of tested amino acid substitutions in conserved residues of the CENH3 histone fold domain lead to uniparental genome elimination on outcrossing
We and others have previously demonstrated that point mutations in some conserved residues of CENH3 induce uniparental genome elimination on outcrossing, leading to haploids that carry only the genome of the wild‐type parent (Karimi‐Ashtiyani et al., 2015; Kuppu et al., 2015). Here, we test additional residues with the goal of identifying all possible HI‐inducing mutations at ethyl methanesulphonate (EMS)‐mutable sites that are conserved among crop species. Since the tail region is extremely variable (Cooper and Henikoff, 2004), even within angiosperms (Maheshwari et al., 2015), we focused exclusively on amino acid residues in histone fold domain (HFD). Aligning the HFD from three different dicot species (A. thaliana, Solanum lycopersicus, B. rapa) and one monocot species (Zea mays), we had previously found that 47 EMS inducible mutations (G to A or C to T transition) would result in same amino acid substitutions in all four species. In our earlier publication (Kuppu et al., 2015), we characterized seven of these amino acid substitutions for their ability, as transgenes, to complement an endogenous null allele and induce haploids upon outcrossing with Ler gl‐1 carrying wild‐type CENH3. Here, we have generated and characterized the effects of thirty‐one additional single amino acid substitutions. Among these new substitution alleles, twenty‐seven resulted in fertile plants with qualitatively normal development, size, and growth rates, as previously observed for seven CENH3 amino acid substitution mutants published earlier. Among these 27 transgenically complemented plants, 19 induced haploids on outcrossing by wild‐type pollen at frequencies ranging from 1.1% to 44% (haploids among surviving seeds, Figure 1a, Table S1, Figure S1). Table 1 lists substitutions that induce haploids among >5% of progeny (a complete list of mutants, including those previously published, is provided in Table S1). In the current study, only a single transgenic line representing each point mutation was tested for HI. It is possible that differences in the level of expression of different CENH3 transgenic lines carrying the same allele might result in changes in haploid induction frequencies. However, in our previous study, independently derived transgenic lines displayed similar levels of HI (these results are reproduced in Table S1), and no HI was observed in three independently derived lines carrying a transgenic wild‐type allele (Kuppu et al., 2015) in a screen of 605 outcrossed progeny. However, the possible effects of differences in expression of different transgenic alleles should certainly be kept in mind if the reader attempts to draw conclusions based on subtle differences in HI frequency.
Figure 1.
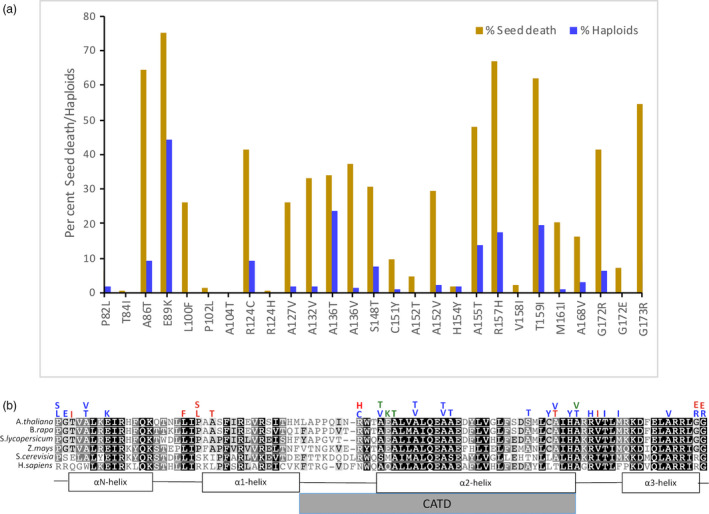
(a) Seed death and haploid induction upon outcrossing. Plants null for the endogenous cenh3 allele carrying a transgene with single amino acid substitutions in the CENH3 histone fold domain were pollinated by wild‐type CENH3 plants carrying the gl1 mutation. Seeds from the cross were assessed for seed death. Per cent haploid induction was calculated as the number of glabrous progeny among all live progeny. (b) Haploid induction rin plants carrying single amino acid substitutions in conserved residues of the CENH3 histone fold domain. Alignment of CENH3 HFD from three different dicot species and a monocot. S. cerevisiae and human CENH3 (CENP‐A) are included to highlight the level of conservation across kingdoms. The coloured letters above the alignment represent single amino acid substitutions tested at that position. Blue represents haploid inducers, red represents substitutions that complement the null allele but do not induce haploids, and green represents the single amino acid substitutions that fail to complement the null allele (lethal alleles). CATD is the CENP‐A (human CENH3) targeting domain. The following mutants were published earlier: P82S, G83E, A86V, P102S, A132T, A137T and G173E (Kuppu et al, 2015). Position of the αN helix, beginning with TVAL, is based on the human H3.1 nucleosome crystal structure.
Table 1.
Tested single amino acid substitutions in the CENH3 histone fold domain that lead to haploid induction at a frequency of 5% or higher
| Codon change | AA change | % Seed death | # Haploids | # Viable progeny | % Haploids |
|---|---|---|---|---|---|
| GGA → GAG | G83E | 36 | 20 | 164 | 12.2 |
| GCT → ACT | A86T | 64 | 4 | 43 | 9.3 |
| GAG → AAG | E89K | 75 | 15 | 34 | 44.1 |
| CGT → TGT | R124C | 42 | 5 | 53 | 9.4 |
| GCG → ACG | A136T | 34 | 5 | 21 | 23.8 |
| TCA → TTA | S148T | 31 | 12 | 155 | 7.7 |
| GCA → ACA | A155T | 48 | 23 | 168 | 13.7 |
| CGT → CAT | R157H | 67 | 6 | 34 | 17.6 |
| ACT → ATT | T159I | 62 | 19 | 97 | 19.6 |
| GGA → AGA | G172R | 41 | 4 | 64 | 6.3 |
| GGA → AGA | G173R | 55 | 9 | 96 | 9.4 |
Plants carrying the mutant allele as a transgene and null for the endogenous gene were pollinated by a CENH3 wild‐type Ler gl1‐1 pollen donor. Haploids were scored using the glabrous phenotype, and % is among viable offspring. Mutant G83E was previously published (Kuppu et al., 2015).
Figure 1b shows the positions of our single amino acid substitutions, with alignment of CENH3 histone fold domain among crop species, and extending the alignment to the CENH3 homologs of fungal (Saccharomyces cerevisiae) and animal (H. sapiens) species. Note that while all 27 of our target amino acids are (by definition) conserved among these four angiosperms, only a subset (perhaps half) tend to be conserved in other kingdoms (Figure S2).
Some conserved residues of the CATD are required for mitotic CENH3 function
Although one would predict that amino acid residues that are highly conserved are required for protein function through roles in catalysis, substrate binding (neither relevant here), localization, folding or interactions with other proteins and nucleic acids, our studies, which have focused exclusively on conserved amino acids, suggest that both the mitotic and meiotic roles of CENH3 are adequately served by mutant proteins carrying a variety of amino acid substitutions in highly conserved residues. However, the protein is not entirely mutable—four different single amino acid changes (among 27 angiosperm‐conserved sites) failed to yield segregants homozygous for the endogenous cenh3‐1 allele, in spite of the fact that several (4–8) independently derived transgenic lines were tested for each construct. This indicates that CENH3 variants carrying these substitutions are nonfunctional (in A. thaliana). A CENH3/cenh3 heterozygote carrying a non‐complementing CENH3 transgene would be expected to display 25% seed death (embryonic lethality) with 1 : 2 segregation for the endogenous CENH3 homozygotes:heterozygotes upon self‐pollination, the same ratio that is observed for a simple heterozygote at the endogenous locus carrying no transgene (Ravi et al., 2010). These ‘non‐complementing’ transgenic lines did segregate at a roughly 1 : 2 ratio (Table 2). We used seed death upon self‐pollination as a second method to determine whether the transgenic alleles were non‐complementing. Plants that are heterozygous for the endogenous CENH3 null (CENH3/cenh3) carrying a single copy of a functional unlinked transgenic point mutant (tgCENH3*) should display seed death of only 6.25% upon selfing, while a CENH3/cenh3 heterozygote exhibits 25% seed death (Ravi et al., 2010). Lines carrying mutations A127T, E128K, A129T and A155V upon selfing had 36.1%, 40.5%, 31.4% and 45.53% seed death, respectively (Table S2), significantly higher than that expected for either a complementing but heterozygous transgene or a simple non‐complementing transgene. This higher rate of seed death suggests that these four transgenic alleles could have a dominant and deleterious effect on reproduction. The 1 : 2 segregation ratio for CENH3 WT:heterozygotes among the surviving seeds suggests that this effect occurs in the sporophyte, not the gametophyte, as both CENH3 and cenh3 alleles are equally transmitted to the zygote (death, and loss of the cenh3 homozygotes, occurs later, during embryogenesis). Although substitutions were tested throughout the angiosperm‐conserved residues of the histone fold domain, these four lethal mutations were found in or adjacent to the alpha‐2 helix, in the more widely conserved CENP‐A targeting domain (CATD), a region required for, and to some degree sufficient for, centromere establishment, maintenance and kinetochore function in yeasts and mammals when swapped into other H3s (Black et al., 2007; Fachinetti et al., 2013; Logsdon et al., 2015; Figure 1b).
Table 2.
Genotypic analysis of non‐complementing point mutations
| Substitution | Independently derived transgenic line | Genotype at endogenous CENH3 (among progeny of heterozygote) | Total progeny genotyped | ||
|---|---|---|---|---|---|
| wt(+/+) | het(+/−) | homo(−/−) | |||
| A127T | M16‐01 | 5 | 7 | 0 | 12 |
| A127T | M16‐03 | 3 | 9 | 0 | 12 |
| A127T | M16‐07 | 9 | 31 | 0 | 40 |
| A127T | M16‐10 | 4 | 8 | 0 | 12 |
| A127T | M16‐11 | 5 | 5 | 0 | 10 |
| A127T | M16‐16 | 4 | 7 | 0 | 11 |
| Total A127T analysed | 30 | 67 | 0 | 97 | |
| E128K | M18‐1 | 4 | 6 | 0 | 10 |
| E128K | M18‐2 | 3 | 12 | 0 | 15 |
| E128K | M18‐3 | 4 | 8 | 0 | 12 |
| E128K | M18‐4 | 6 | 6 | 0 | 12 |
| E128K | M18‐5 | 9 | 3 | 0 | 12 |
| E128K | M18‐6 | 10 | 13 | 0 | 23 |
| E128K | M18‐7 | 3 | 9 | 0 | 12 |
| Total E128K analysed | 39 | 57 | 0 | 96 | |
| A129T | M19‐02 | 2 | 4 | 0 | 6 |
| A129T | M19‐04 | 11 | 22 | 0 | 33 |
| A129T | M19‐07 | 5 | 5 | 0 | 10 |
| A129T | M19‐09 | 2 | 8 | 0 | 10 |
| A129T | M19‐10 | 7 | 4 | 0 | 11 |
| Total A129T analysed | 27 | 43 | 0 | 70 | |
| A155V | M35‐01 | 8 | 22 | 0 | 30 |
| A155V | M35‐02 | 6 | 18 | 0 | 24 |
| A155V | M35‐03 | 10 | 16 | 0 | 26 |
| A155V | M35‐22 | 3 | 8 | 0 | 11 |
| Total A155V analysed | 27 | 64 | 0 | 91 | |
T2 and T3 seeds of plants that carried the transgenic variant version of CENH3 and were heterozygous for the endogenous CENH3 null allele were grown on selective medium, transplanted to soil and genotyped for endogenous CENH3 locus. For the above four mutations, lines homozygous for the null allele at the endogenous locus were not obtained from among >90 genotyped progeny.
Double amino acid substitutions can increase the frequency of haploid induction
One would predict that a combination of two amino acid changes in a single allele at CENH3 would have a more severe phenotypic effect than either single amino acid change alone. We tested six combinations of mutations (Table S3), combining two non‐inducers, two high inducers, and one high and one low inducer. The double mutant versions of the histone fold domain were synthesized and cloned into destination vector as described in Methods. Of the six double mutant combinations tested, all were able to complement the endogenous cenh3 null mutation. Five out of the six combinations tested resulted in haploid induction at a rate equal to or higher than the average of two independent single amino acid substitutions (Figure 2 and Table S3). The combination of P82L and A132T showed a sixfold increase in haploid induction rate over the average of the two different single amino acid substitutions. It is also useful to note that combining two non‐inducer point mutations resulted in an HI line. Interestingly, one combination of alleles [G83E (HI:11%) and G172R (HI:6.3%)] produced haploids at a lower frequency (2.3%) than either parent indicating positive intramolecular epistasis.
Figure 2.

Haploid induction by double mutation. Double mutants in an endogenous cenh3 null background were pollinated by wild‐type CENH3 plants carrying the gl1 mutation. Percent haploid induction was calculated as the number of glabrous progeny among all live progeny.
In‐frame deletion mutations in the CENH3 histone fold domain lead to haploid induction
EMS‐inducible amino acid changes were chosen for this study as these mutations are currently unregulated. However, these mutants must be isolated through random mutagenesis, which requires the screening of large populations of plants. CRISPR/Cas9‐generated mutants are unregulated in some countries, though simple amino acid substitutions are a relatively rare CRISPR product, the majority of mutations being deletions or single base additions. However, in an effort to generate a null allele in tomato we found that we recovered a high frequency of in‐frame deletions at the target site—the beginning of the HFD. The vector used to transform tomato cotyledons with Agrobacterium rhizogenescarried a T‐DNA encoding the 2x35SΩ:Cas9, AtU6‐26 driving the single guide RNA, and citrine–tailswap was driven by the SlCENH3 native promoter (Figure 3a). On transformation, both the R‐DNA and the engineered T‐DNA, carrying citrine–tailswap, a selectable marker, and CRISPR/Cas9 targeting the beginning of the CENH3 αN helix, stably integrate into the genome and initiate the formation of hairy roots in tomatoes (Bulgakov, 2008; Ron et al., 2014). The emerging roots were assayed for gene editing in the endogenous CENH3 locus. In hairy roots obtained from two independent experiments, we characterized 11 independently derived lines where the PCR‐amplified endogenous CENH3 alleles were resistant to digestion by the restriction enzyme BsmF1 (recognition site GGGAC), indicative of mutation at the target. The PCR‐amplified CENH3 target region from eleven BsmF1‐resistant roots was sequenced. Seven out of the eleven characterized mutant roots were apparently homozygous for the gene editing (though some alleles may be resistant to amplification). The remaining 4 were biallelic (Table 3, Figure 3b). Nine out of the eleven characterized mutants had in‐frame deletions in CENH3 locus either in the homozygous (7/10) or biallelic (3/10) state—a surprisingly high fraction of in‐frame deletion mutants. There was also one root with an out of frame (7 bp) homozygous deletion, and a second with a larger 152 bp homozygous deletion that spanned an intron/exon junction (the nature of the resulting splice product remains undetermined). This apparent overrepresentation of in‐frame alleles raised the question of whether the GFP–tailswap roots carrying the in‐frame deletions are viable because these mutations are functional CENH3 alleles—and perhaps the citrine–tailswaptransgene is only weakly supporting CENH3 function.
Figure 3.
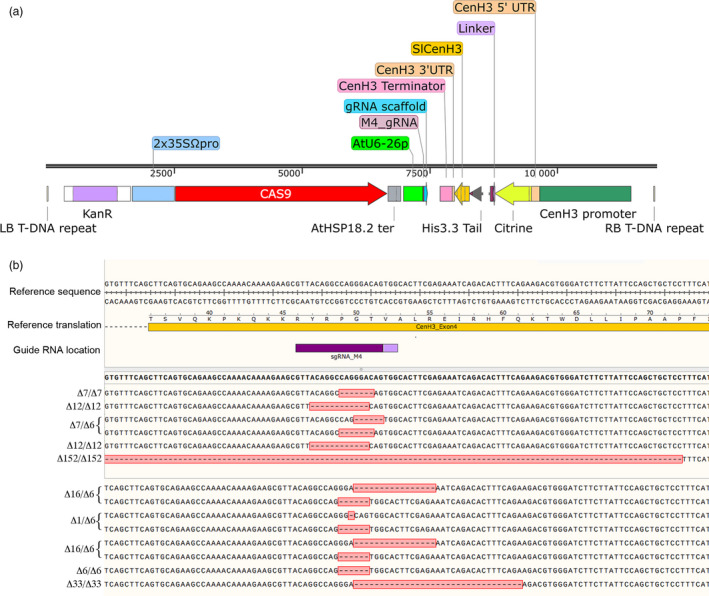
(a) Map of the region between the left border and right border of the vector used for transformation of tomato cotyledons with Agrobacterium rhizogenes. The expression of cas9 was driven by 2x35S promoter, and guide RNA was driven by AtU6‐26 promoter. Citrine–tailswap is N‐terminal fusion of citrine to the Solanum lycopersicum histone 3.3 N‐terminal tail, fused to the histone fold domain of SlCENH3. The native SlCENH3 promoter was used to drive the expression of citrine–tailswap. (b) DNA preps from hairy roots were screened for mutant alleles via BmsF1 resistance; resistant alleles were cloned and sequenced. Reference sequence and location of the guide RNA are shown in the top. BsmF1 recognition site = GGGAC.
Table 3.
Genome editing at the endogenous CENH3 locus in hairy root lines of tomato
| Line # | Deletions (bp) | In‐frame deletion present |
|---|---|---|
| 1 | ∆7/∆7 | No |
| 2 | ∆12/∆12 | Yes |
| 3 | ∆7/∆6 | Yes |
| 4 | ∆12/∆12 | Yes |
| 5 | ∆152/∆152 | Unknown |
| 6 | ∆16/∆6 | Yes |
| 7 | ∆1/∆6 | Yes |
| 8 | ∆6/6 | Yes |
| 9 | ∆16/∆6 | Yes |
| 10 | ∆6/∆6 | Yes |
| 11 | ∆33/∆33 | Yes |
The guide RNA was designed to target the beginning of exon 4, at the beginning of the histone fold domain. Genomic DNA was extracted from independently derived hairy roots, and the endogenous CENH3 locus was amplified used primers flanking the gRNA target. PCR products that were not digested by the restriction enzyme BsmF1 (which recognizes the wt allele) were cloned into a plasmid vector and sequenced.
To determine whether in‐frame deletions could complement an endogenous null mutation, we generated two and eleven amino acid deletions in the A. thaliana CENH3 histone fold domain that mimic the deletions obtained in the tomato hairy root assay. These transgenic constructs were transformed into CENH3+/− A. thaliana. In the T1 generation, we found several cenh3−/− plants that were complemented by Δ6(2aa) deletion allele and, remarkably, the Δ33bp(11aa) deletion in the αN helix of CENH3. We used the online tool Phyre (Kelley et al., 2015) to predict the structure of A. thaliana wild‐type CENH3. This predicted structure (Figure S3) suggests that the 11 aa deletion encompasses nearly all of the highly conserved αN helix. It should also be noted that plants carrying these transgenic deletion alleles, and null for the endogenous allele, grow normally and are fully fertile, indicating that this helix is not required for CENH3 mitotic or meiotic function in A. thaliana.
To determine whether these complemented lines were haploid inducers, we pollinated them by Ler gl‐1. Two independent transgenic lines carrying the two amino acid deletion (Δ2–7 and Δ2–12) produced haploids at a frequency of 25.7% and 8.0%, respectively. Two independent lines carrying the eleven amino acid deletion (Δ11–8 and Δ11–18) produced haploids at a frequency of 16% and 17%, respectively (Table 4).
Table 4.
Deletion mutations in the αN helix of the CENH3 histone fold domain result in haploid induction on outcrossing
| Mutation | % Seed death | # Of haploids | Total # of viable offspring | % Haploids |
|---|---|---|---|---|
| △6 line 7 | 70 | 18 | 70 | 26 |
| △6 line 2 | 78 | 2 | 23 | 8 |
| △33 line 8 | 72 | 4 | 25 | 16 |
| △33 line 18 | 68 | 14 | 83 | 17 |
Plants carrying the mutant allele transgenically and null for the endogenous gene were pollinated by a CENH3 wild‐type Ler gl1‐1 pollen donor. Haploids were scored using the glabrous phenotype as a proxy. Haploids were scored using the glabrous phenotype, and % is among viable offspring.
Seed death on outcrossing by wild‐type CENH3 is correlated with haploid induction
Plant embryogenesis is marked by a double fertilization event, in which the pollen tube delivers two sperm nuclei. One nucleus fuses with the egg to form the zygote, while the other fuses with the diploid central cell to form the triploid endosperm, a tissue that supports embryonic growth. Although genome elimination is assayed in seedlings, it can also occur in the endosperm (Ravi et al., 2014). In addition, outcrossed haploid inducers often produce aneuploid progeny among the surviving F1 (Ravi and Chan, 2010) and there may be more severely aneuploid embryos that spontaneously abort during development. Endosperm failure due to aneuploidy or imbalances between the male‐ and female‐derived genomes can also occur. An outcross performed with a strong haploid inducer would then be expected to result in high frequencies of aneuploidy (in embryo and the endosperm) and possibly haploid (vs. normal triploid) endosperm, any of which might result in seed abortion. As illustrated in Figures 1a and 4, we found that, indeed, there is a strong correlation (r = 0.8) between haploid induction on outcrossing and seed death on outcrossing (again, neither effect occurs on self‐pollination, provided the complementing transgene is homozygous). Seed death is in fact a more severe, more pervasive and more easily observed effect of certain amino acid substitutions (as well as other CENH3 mutations). Thus seed death, observed only on outcrossing, might be employed as a sensitive predictor of HI in potential HI lines.
Figure 4.

Correlation between seed death and haploid induction. Haploid‐inducing mutations including GFP–tailswap, CENH3 from other species, point mutations, double mutations and deletions mutations were pollinated by Ler gl1 wild‐type for CENH3. Percent seed death upon outcrossing was plotted against percent haploid induction. This graph displays data from all known single point mutants, double point mutants, deletion mutants, as well as the endogenous cenh3 null complemented by CENH3 from Lepidium oleraceum and Brassica rapa (Maheshwari et al., 2015) and GFP‐tailswap (Ravi and Chan, 2010).
CENH3 variant proteins are displayed on chromatin in a pattern indistinguishable from wild type
It is possible that our variant CENH3 proteins localize abnormally (to many sites) and/or weakly to the centromere. To test the localization pattern of variant CENH3 protein on the centromere, we performed immunostaining with AtCENH3 antibody (Talbert et al., 2002a) on the nuclei isolated from young seedlings carrying CENH3 variants representing the various classes of mutants [single (R124C, A115T), double (R124C and A115T) and deletion (Δ2–7)] along with the wild‐type CENH3 lines. We were able to detect the localization of variant CENH3 to the centromere in the nuclei isolated from 10‐day‐old seedlings in all of the lines tested (Figure 5a). In both variant and wild‐type lines, at least one‐third of the observed nuclei had ten distinct signals as expected for diploid A. thaliana (Figure 5b). In the nuclei in which chromocentres were prominent, the majority of the signals were detected in the space proximal to the chromocentres, marked by relatively intense DNA staining with DAPI. All nuclei analysed can be seen in Figure S6. Overall, the numerical (number of spots) and qualitative variation (of intensity of spots within a nucleus) of CENH3 signals within a given nucleus and across nuclei within the same mutation displayed a comparable pattern to that found with wild‐type CENH3 signals, consistent with previous observations (Lermontova et al., 2006). Taken together, there was no readily distinguishable variation in localization patterns found among the mutants when compared with the wild type.
Figure 5.
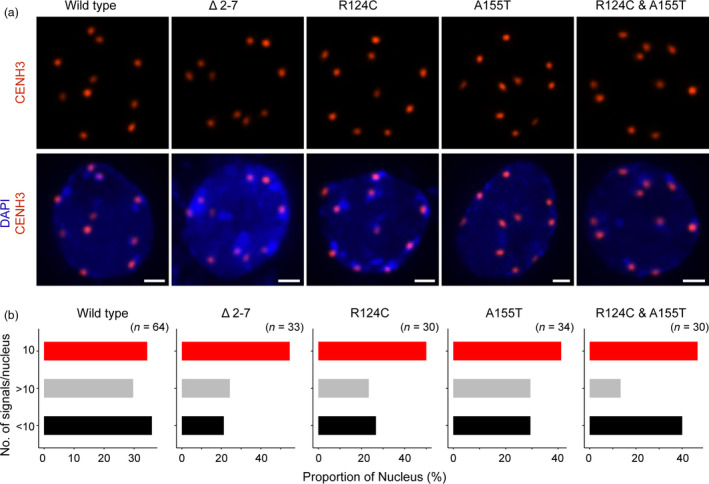
Immunolocalization analysis of wild‐type and variant CENH3 proteins on nuclei isolated from young seedlings. (a) Representative nuclei displaying ten distinct CENH3 signals following maximum intensity projection from all genotypes analysed. All the micrographs were acquired and processed similarly. Scale bars = 1 μm (b) Bar chart displaying CENH3 signals in each nucleus from all genotypes analysed, and the number of nuclei analysed for each genotype is given in the parenthesis.
Point, double and deletion mutations in CENH3 act as a haploid inducers only when the HI line is used as the female/seed parent
In all the mutations described above, the transgenic CENH3* mutant in the background of endogenous cenh3‐1 null was used as a female parent. To test for parent of origin effect on haploid induction, we switched the direction of cross. Here, we used Ler gl‐1 as the female and tgCENH3* mutants in the cenh3‐/‐ background as pollen donors. A comparison of the forward and reciprocal cross is shown in Figure 6a,b, respectively (also Table S4a). When used as seed parent, the mutants P82S, G83E, P102S, A136T, Δ2–7 and the double mutant M4 and M34 induce haploids frequency of 2.4%, 12.2%, 0%, 2.26%, 23.88 and 25.71%, respectively. The same mutants, when used as a pollen parent, do not induce haploids at the scale tested here (Table S4b). It is interesting to note that the seed death upon outcrossing also decreases when the direction of the cross is reversed—but it is not entirely eliminated. This suggests that haploid induction might also be observed when the HI line is used as a male, provided enough progeny could be screened (Ravi and Chan, 2010).
Figure 6.
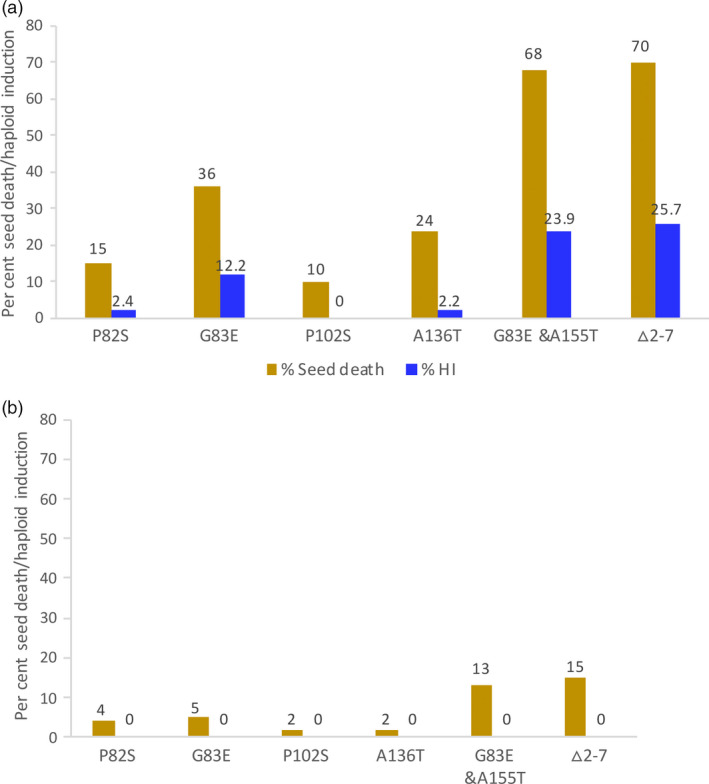
Comparison of haploid induction in the forward vs reciprocal cross. In forward crosses (a), the variant CENH3* transgenics in a cenh3 null background were used as the female parent and wild‐type CENH3 Lergl1 was used as male parent. In reciprocal crosses (b), the wild‐type CENH3 Ler gl1 was used as the female parent and variant CENH3 transgenics in a cenh3 null background were used as the male parent.
Point mutants obtained by tilling CENH3 can induce haploids only in homozygous state and not in heterozygous state with the wild‐type allele
TILLING is a methodology for screening large populations of densely mutagenized plants for mutations induced at a target chromosomal sequence (Comai and Henikoff, 2006). We previously demonstrated that both the transgenic allele A86V (in a null cenh3 background) and the tilling allele A86V can serve as haploid inducers, producing haploids at a frequency of 2.7% (Kuppu et al., 2015). To test whether the A86V tilling mutation would produce haploids when in a heterozygous state with the wild‐type allele, we performed test crosses using the heterozygote (A86V/WT). Seed death was 1.2%, but among the 1021 offspring that germinated none were haploids. Thus, the A86V tilling mutant acts as an efficient haploid inducer only in the absence of the wild‐type CENH3 allele.
Prediction of HI alleles
Our study of amino acid substitutions has focused on EMS‐inducible nucleotide changes that result in the same amino acid substitution in A. thaliana, B. rapa, Saccharomyces esculentum and Z. mays, in the spirit of translatability. As a result, our selected targets tend to be conserved among angiosperms in general. However, when put into practice in an EMS‐induced TILLING population, new alleles will not be restricted to these specific amino acid changes. The researcher who is considering testing HI in these randomly mutagenized stocks may want to obtain an estimate of the significance of their randomly induced amino acid change. This requires consideration not only of position of the amino acid in question, but also the nature of the specific amino acid change. Many programs exist that are intended to predict severity of mutant alleles, taking into consideration, to varying degrees, the nature of the change in R group (sometimes in context of tertiary structure), the frequency at which such changes are found in natural populations of the same species, and evolutionary conservation across kingdoms (Malhis et al., 2019). One such program is SIFT (sorting intolerant from tolerant; Ng and Henikoff, 2003). SIFT takes into account structure, sequence and annotation information among a wide range of species to calculate output, with values ranging from 0 to 1, where zero is ‘not tolerated’ and 1 is ‘tolerated’. Scores below 0.05 are considered predictive of ‘functionally not tolerated’. To see whether there was any correlation between SIFT score vs haploid induction and seed death, we plotted their relationship (Figure 7a,b) and found that among the best haploid inducers—those with 5% or greater HI—all but one had a SIFT score less than 0.05. The exception—G172R, which has an HI rate of 6.3%—is at a residue that is somewhat variable among angiosperms. It is found as a G in most of the wide range of 53 angiosperms we previously checked (Kuppu et al., 2015), and otherwise as S, T or, in Lotus japonicus (trefoil), R itself (Kuppu et al., 2015). R is also found at this site in humans and S. cerevisiae, which explains its very high (‘well tolerated’) SIFT score. This result suggests that although there is a very strong correlation of HI with SIFT score, there are exceptions, and amino acid substitutions in the HFD that are not observed within closely related species have the potential to be haploid inducers, regardless of SIFT score.
Figure 7.
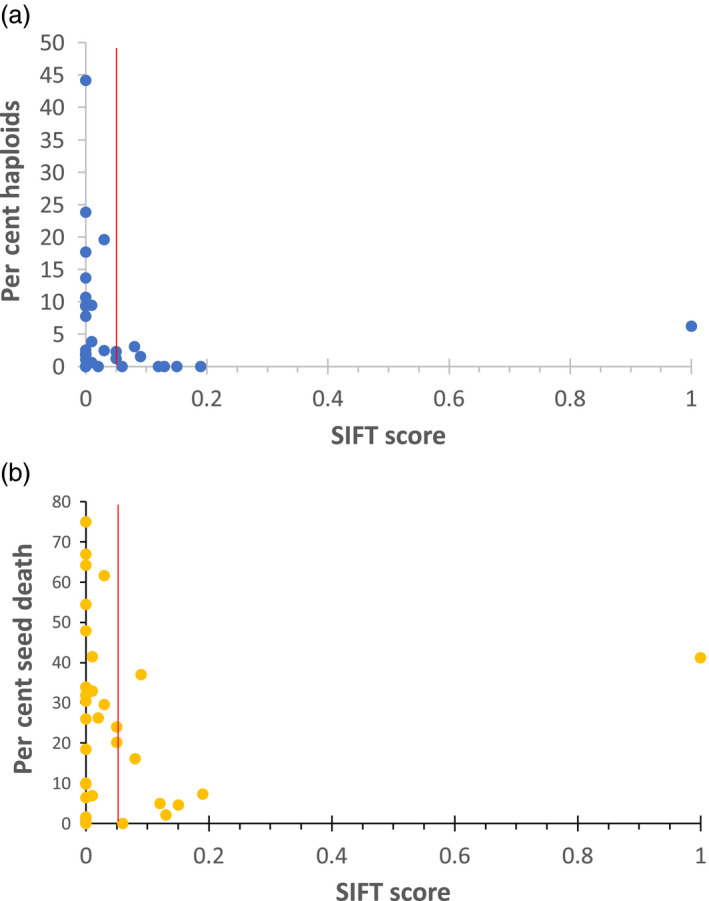
Substitution of highly conserved amino acids under purifying selection leads to genome elimination. Plot of SIFT (sorting intolerant from tolerant) scores against percent haploid induction (a) and percent seed death (b). The red line indicates the SIFT score of 0.05. Amino acid substitutions with a SIFT score of 0.05 are generally predicted to be intolerant and the substitutions with a score of 0.05 and higher are predicted to be tolerated.
Discussion
The root cause of CENH3‐induced HI remains unknown. CENH3 has two core and perhaps separable functions: to attract CENH3/H4 reloading factors to the site of existing CENH3 nucleosomes, and to act as a platform for the building kinetochores. It has been proposed (Chan, 2010; Comai et al., 2017; Ishii et al., 2016; Sanei et al., 2011; Wang and Dawe, 2018) that there is some difference in either the quality (recognizability of CENH3* vs. CENH3 nucleosomes by CENH3/H4 loading or more distal centromere components) or quantity (number of CENH3 nucleosomes per centromere, the root cause of which must be based on the quality of the CENH3* allele) between the centromeres carried by the gametes derived from mutant and wild‐type parents. This difference in quality or quantity results in a delay in reloading and/or kinetochore building on the CENH3* centromeres when the two sets of chromosomes are placed in competition for these maintenance and/or mitotic factors, resulting in the functionalization of the centromeres on the wild‐type, but not the mutant, chromosomes. The inert mutant‐derived chromosomes (or some of them, in aneuploids) are then left behind at the metaphase plate, and sometimes reincorporated into the nucleus as extensively rearranged chromosomes (Tan et al., 2015). The mitotic checkpoint may be ignored, if the mutant‐derived centromeres have not attracted enough kinetochore components to be recognized as such. In contrast, in early embryogenesis after self‐pollination, all centromeres are equal, all are presumably reloaded with CENH3 or more distal components at similar rates, and the cell cycle is unperturbed. This may explain wild‐type growth and fertility observed here and elsewhere in even extreme HI inducers, such as E89K (44% HI).
Ravi and Chan serendipitously discovered CENH3‐mediated haploid induction in 2010 (Ravi and Chan, 2010). Their method involved knocking out the endogenous CENH3 and expressing a chimeric CENH3 (‘GFP–tailswap’). Since then, other modifications of CENH3 have been shown to induce haploids in A. thaliana. Complementation of the cenh3 null with CENH3 from related species (B. rapa, L. oleraceum) leads to genome elimination on outcrossing with plants carrying wild‐type CENH3 (Maheshwari et al., 2015). Amino acid substitutions in the histone fold domain of CENH3 lead to haploid induction as well (Karimi‐Ashtiyani et al., 2015; Kuppu et al., 2015). CRISPR‐induced in‐frame deletions also create excellent haploid inducers (this paper). Given that CENH3 performs the same function in all eukaryotes, this would appear to be a promising candidate for translation to other species. However, in the 9 years since Ravi and Chan’s first publication, there has been only one reported instance of a (non‐Arabidopsis) HI line produced by modification of CENH3, in maize (Kelliher et al., 2016). This experiment involved a ‘GFP–tailswap’ equivalent allele, in a background carrying a null mutation of the native CENH3. HI was observed, though at a relatively low frequency vs. the native stock 6 induction system or the A. thaliana GFP:tailswap line. Recently, a CENH3 allele that mimics a known HI inducer of A. thaliana (Karimi‐Ashtiyani et al., 2015) was discovered in a chickory species (Cichorium intybus), and may be linked to this line’s HI‐inducing properties when crossed, as a female, by a related species (Cicerbita alpina) carrying the wild‐type allele (vs. A. thaliana) at this same CENH3 ortholog (Van der Veken et al., 2019). It is possible that one or more aspects of CENH3 biology may vary between plant species and that A. thaliana is, for unknown reasons, highly tolerant of changes in CENH3, in terms of mitotic and meiotic function, while being sensitive to quantitative or qualitative differences between parental gametes. The study of GFP–tailswap in maize suggests there may be differences between angiosperms in the CENH3 biology that produces haploids—Keliher et al. (2016) observed a higher rate of HI when the HI line was used as the pollen donor, in dramatic contrast to our result here, in which we observe HI only when the HI line is used as a the female.
Here, we demonstrate that in‐frame deletions can exhibit partial loss of function while maintaining viability and fertility on self‐pollination. Specific amino acid substitutions can also be generated in crops amenable to CRISPR/Cas9 mutagenesis, via cas9‐guided base editors (Marzec and Hensel, 2018). Our results suggest that CENH3 can tolerate multiple simultaneous edits. It is hoped that these easily accessed CRISPR‐based technologies will stimulate investigation into the effects of CENH3 partial loss‐of‐function mutations in a wider variety of crops.
Given our exclusive focus on amino acids that are highly conserved among angiosperms, and often conserved within eukaryotes, it was interesting to find that among a total of 38 substitutions characterized, only four lethal single amino acid substitutions were identified: A127T, E128K, A129T and A155V. All of these amino acids are located in the CENP‐A (human CENH3) Targeting Domain (CATD). This region is particularly well‐conserved between kingdoms (Figure S2) and is thought to be required for both the timing of deposition and localization to the centromere. In humans, chimeric histone H3s that contain the CATD (loop 1 and α2 helix from CENP‐A) were able to assemble into human centromeric chromatin (Black et al., 2004).
All other substitutions resulted in healthy and fertile plants. Similar results regarding the flexibility of the majority of amino acids in CENH3 have previously been obtained in budding yeast (Camahort et al., 2009). In this thorough study of the CENH3 homolog CSE4, in which every amino acid was systematically changed to alanine, only six of these substitutions were found to be lethal in yeast. All six of these sites are widely conserved among eukaryotes (Figure S2)—they are among the only 19 amino acids conserved among humans, A. thaliana, Drosophila and budding yeast. None of these sites match our four lethal substitutions. Three discrepancies are easily explained: among our lethal substitutions, three involve a highly conserved alanine, and so were not tested in the yeast study. Our fourth lethal allele is at a site that is extremely conserved among angiosperms (E in 51/53 angiosperms checked, Figure S4), but not among eukaryotes. The critical nature of this site (E128) may be angiosperm‐specific, perhaps due to interactions with loading or kinetochore‐building factors that are angiosperm‐specific. Among the lethal substitutions observed in yeast,we have tested substitutions at two positions, H154Y (A. thaliana numbering) and R157H. Both substitutions are viable in A. thaliana (though haploid‐inducing). This might be due to a particular sensitivity in S. cerevisiae to substitutions at this site, or because our substitutions are more conservative changes than the substitution to an alanine studied in budding yeast.
Our findings that double mutants (two amino acid substitutions in a single protein) generally can induce haploids at a frequency higher than that of the single mutants are not surprising and indicate that synergistic epistasis is possible. This presents an opportunity to ‘improve’ a weak TILLING‐derived HI line through an additional round of EMS mutagenesis. The fact that double mutants are tolerated is also encouraging from the perspective of deploying base‐editor forms of CRISPR to generate targeted amino acid changes. Although these editors can be guided by an inactivated short guide/Cas9, they are not an intrinsic part of Cas9 but are instead artificially tethered to Cas9, resulting in a window, rather than a unique site, of base modification (Marzec and Hensel, 2018). However, our results indicate that CENH3 can tolerate multiple simultaneous edits, and these multiply‐edited mutations may be useful. The fact that in‐frame deletions in the CENH3 histone fold domain are able to induce haploids presents a new and simple approach to the generation of haploid inducers, eliminating the transgenic GFP–tailswap approach (which also requires isolation of a null allele at this essential gene), or the production of large TILLING populations.
From the immunostaining analysis of the wild‐type and variant CENH3‐expressing plants, we were able to visualize the expected 10 signals and their association with chromocentres (when chromocentres were identified), consistent with the pattern of localization observed in wild type. The variation in signal intensity observed in nuclei isolated from wild‐type seedlings was comparable to that observed in the mutant lines. The fact that none of the mutants (endogenous null kept alive by a variant version of CENH3) show any obvious vegetative or reproductive phenotype is consistent with this wild‐type localization pattern.
In our previous study (Kuppu et al., 2015), we analysed the anthers of CENH3 variant mutants by Alexander staining and did not see any decrease in pollen viability, suggesting that there is no significant aneuploidy in the meiotic products. Another related study (Maheshwari et al., 2015) found that when A. thaliana CENH3 is replaced with the CENH3 from Z. mays, ChIP‐Seq analysis displayed a pattern of binding to DNA sequences that was indistinguishable from that of native CENH3. Further, these hetero‐complemented A. thaliana plants display normal growth and are fully fertile upon selfing, but result in genome elimination upon outcrossing—the same phenotype by our point mutants, double mutants and deletion mutants.
We have generated a 3D model of AtCENH3 in the context of nucleosome complex (Figure 8a), using homology modelling based on a crystal structure of a canonical nucleosome (Tachiwana et al., 2011b). Modelling of all 31 mutations (positions shown in Figure 8b) resulted in no predicted changes in the overall complex architecture (Figure 8b,c). The eleven amino acid deletion, encompassing nearly all of the αN helix, includes three positively charged amino acids (TVALKEIRHFQ), preceded by a high density of positive charge (SQKKSYRYRPG; Figure 8c, Figure S3), which is highly conserved (in terms of charge) among eukaryotes (Figure S5).
Figure 8.
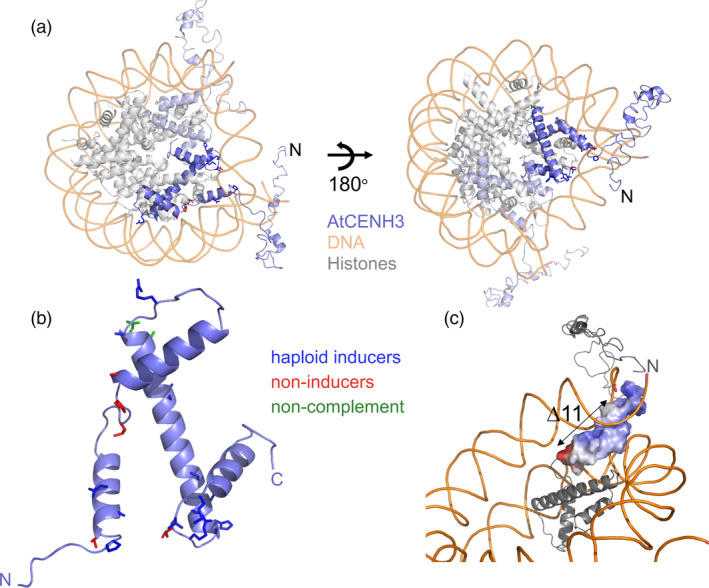
Predicted structure of Arabidopsis CENH3 with sites of mutations. (a) AtCENH3 predicted structure aligned to nucleosome 3av1 (Tachiwana et al., 2011b), a human nucleosome carrying histone 3.2. (b) AtCENH3 with side chains indicating the locations of the mutations: blue = haploid inducers, red = non‐inducers, green = lethal. (c) Electrostatic charge of the alphaN helix (TVALKEIRHFQK) and the immediately adjacent 11 amino acids of the N‐terminal tail region (SQKKSYRYRPG) are displayed as a space‐filling model. Blue indicates positively charged and red negatively charged.
Given the trans‐kingdom conservation of this helix among canonical H3s, H3.3s and all angiosperm CENH3s checked (consensus among H3s = TVALREIRRYQK; Figures S4 and S5), it was surprising that virtually the entire αN helix (Figure S3) can be deleted and a haploid‐inducing (but mitotically and meiotically functional) allele could be obtained. The H3 αN helix and the positively charged loop region that precedes it are thought to stabilize and orient the exit of the DNA from the nucleosome (Luger et al., 1997; Tachiwana et al., 2008; Tsunaka et al., 2005). However, this region is poorly conserved in many if not most animal, fungal and protozoan CENH3 homologs (Figure S5). The crystal structure of human CENP‐A in a nucleosomal context (Tachiwana et al., 2011a) depicts an abbreviated αN helix, rather than a helix of conserved length but diverged sequence. This abbreviated helix appears to destabilize the interaction with DNA at the exit site (significantly less DNA is visible) vs. a conventional H3. The flexibility of the length of the αN helix among eukaryotic CENH3 homologs might indicate that this region plays a role in CENH3‐specific (rather than more routine nucleosomal) functions, which in turn are governed by rapidly evolving interacting proteins. If so, we can conclude that this surface for interaction with maintenance and/or kinetochore‐building proteins is reasonably conserved among angiosperms, and these defects in these CENH3:DNA interactions have a large effect on ‘competition’ between chromosomes loaded with variant CENH3 and wild‐type CENH3 at the centromeres in the zygote.
Here, we summarize a variety of approaches for creating haploids by CENH3‐mediated haploid induction. Single amino acid substitutions in CENH3 histone fold domain can produce haploids at frequency as high as 44%. Combining two single amino acid generally increased the frequency of haploid induction. Our latest finding that simple CRISPR/Cas9‐mediated deletions in the αN helix can induce haploids opens a new avenue for the generation of haploid‐inducing lines. The technology is available to generate similar mutations in crop plants, but it remains to be seen whether the remarkable effects of these mutations—producing vigorous, self‐fertile plants that are also efficient haploid inducers—can be translated to crop species.
Methods
Plant growth conditions
Arabidopsis thaliana plants of ecotypes Col and Ler were grown in a Conviron growth chamber under following conditions: 16‐h light/8‐h dark cycle, 20 °C, 55% relative humidity. Sunshine Mix #1 Fafard 1P (Sungro Horticulture, Agawam, MA) was used to grow the plants.
Cloning CENH3 mutants
pCAMBIA 1300 was used as a base vector for our constructs. Destination vector for LR recombination to introduce single point mutant, double point mutant and deletion mutant was created by 4‐step cloning. Step 1, we amplified the promoter (1 Kb upstream of ATG) and cloned into pCAMBIA 1300. Step 2, we cloned the terminator and the 3′ UTR region of CENH3. Step 3, in the multiple cloning site between the promoter and the terminator region between the KpnI site and XbaI sites, the N‐terminal tail region of the CENH3 with introns until first half of intron 5 was cloned. Ccdb cassette with attR1 and R2 sites with and chloramphenicol resistance gene was cloned between the CENH3 tail and the 3′UTR region using the restriction sites Bgl1 and XbaI. The point mutants, double mutants and the deletion mutants were synthesized with flanking L1 and L2 sites from Genewiz (Genewiz, NJ). The synthesized fragment was LR recombined (a recombination between attL sites and attR sites; Gateway Cloning Technology, Invitrogen‐Thermo Fischer Scientific, Waltham, MA, USA) with destination vector to obtain the expression vector. The expression vectors were sequenced and transformed into Agrobacterium tumefaciens strain GV3101.
Construction of the citrine–tailswap/cas9/sgM4 vector
A detailed method for these sections is included in Methods S1.
DNA extraction and genotyping
Protocol for this section has been included in Methods S1.
Crossing scheme and haploid screening
Methodology for this section is provided under Methods S1.
Immunostaining and imaging
Methodology for this section is provided under Methods S1.
Structural prediction and analysis
The predicted model structure of AtCENH3 was generated and analysed using web‐based homology modelling server Phyre2 (http://www.sbg.bio.ic.ac.uk/∼phyre/) and I‐TASSER (https://zhanglab.ccmb.med.umich.edu/I‐TASSER/) with 0.41 ± 0.14 estimated TM score. Based on BLAST‐PDB (NCBI) analysis with the highest total score and identity (112 and 47.48% respectively), AtCENH3 predicted structure was aligned (RMSD = 0.154) with histone H3.2 in the context of human nucleosome crystal structure (PDB: 3AV1; Tachiwana et al., 2011b). Analyses, alignments and graphical representations were generated with PyMOL (version 2.4.0). Electrostatics presentation was generated in PyMOL via vacuum electrostatics with range ±100.0.
Conflict of interest
A patent application has been submitted by SK, ABB and MR for the induction of HI by CRISPR as demonstrated in this work.
Author contributions
SK and ABB conceived the idea and designed the experiments with inputs from LC. SK, MR, MPAM, GL, AH, MHS, JT and RB performed the experiments. SK, MR, MPAM, GL and ABB analysed the data. NS and SK did the web‐based homology modelling of AtCENH3 protein/nucleosome structure. SK and ABB wrote the paper with inputs from MR, MPAM, NS and LC.
Supporting information
Figure S1 Map of CENH3 histone fold domain showing amino acid substitutions and their haploid inducing frequency.
Figure S2 Alignment of D. melanogaster, S. cerevisiae, Arabidopsis thaliana, and H. sapiens CENH3 homologs.
Figure S3 Predicted structure of Arabidopsis thaliana CenH3 showing the regions of 2 and 11 amino acid deletions.
Figure S4 Sequence alignment of the HFD of CENH3 from 53 different angiosperm species.
Figure S5 Illustrating the conservation across eukaryotes of the alphaN helix among conventional H3’s, but not CENH3 (Draizen et al., 2016).
Figure S6 Pooled data from immunolocalization analysis of wild‐type and variant CENH3 proteins on the nucleus isolated from 10 days old seedlings.
Table S1 Showing all the point mutants tested so far for their ability to complement endogenous CENH3 null, set seeds and induce haploid upon out crossing to Ler gl1 carrying wild‐type CENH3
Table S2 Assessing seed death upon selfing for non‐complementing mutation
Table S3 Haploid induction frequencies of double point mutants tested
Table S4 Percent seed death and haploid induction rate of crosses where the variant CENH3 complementing endogenous cenh3 null was used either as a female (a) or a male (b) parent
Methods S1 This is a diagram showing where we chose to mutate amino acids, rather than an experimental result it summarizes what is described in the paper
Acknowledgements
We would like to thank Steven Henikoff and Paul B. Talbert for sharing generous aliquots of the CENH3 antibody. We would also like to thank MCB Light Microscopy Imaging Facility in UC Davis for providing infrastructure for immunofluorescence imaging and analysis. We thank Dr. Harmit S Malik (Fred Hutchinson Cancer Research Center, Seattle, WA) for his input on the analysis of SIFT scores and haploid induction. We also thank Rijk Zwaan Zaadteelt en Zaadhandel B.V., East‐West Seed International and Syngenta for their continued support of this project.
Kuppu, S. , Ron, M. , Marimuthu, M. P. A. , Li, G. , Huddleson, A. , Siddeek, M. H. , Terry, J. , Buchner, R. , Shabek, N. , Comai, L. and Britt, A. B. (2020) A variety of changes, including CRISPR/Cas9‐mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol. J., 10.1111/pbi.13365
References
- Allshire, R.C. and Karpen, G.H. (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9, 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B.E. , Foltz, D.R. , Chakravarthy, S. , Luger, K. , Woods, V.L. Jr and Cleveland, D.W. (2004) Structural determinants for generating centromeric chromatin. Nature, 430, 578. [DOI] [PubMed] [Google Scholar]
- Black, B.E. , Jansen, L.E. , Maddox, P.S. , Foltz, D.R. , Desai, A.B. , Shah, J.V. and Cleveland, D.W. (2007) Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP‐A targeting domain. Mol. Cell, 25, 309–322. [DOI] [PubMed] [Google Scholar]
- Bulgakov, V.P. (2008) Functions of rol genes in plant secondary metabolism. Biotechnol. Adv. 26, 318–324. [DOI] [PubMed] [Google Scholar]
- Camahort, R. , Shivaraju, M. , Mattingly, M. , Li, B. , Nakanishi, S. , Zhu, D. , Shilatifard, A. et al (2009) Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell, 35, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W. (2010) Chromosome engineering: power tools for plant genetics. Trends Biotechnol. 28, 605–610. [DOI] [PubMed] [Google Scholar]
- Comai, L. and Henikoff, S. (2006) TILLING: practical single‐nucleotide mutation discovery. Plant J. 45, 684–694. [DOI] [PubMed] [Google Scholar]
- Comai, L. , Maheshwari, S. and Marimuthu, M.P. (2017) Plant centromeres. Curr. Opin. Plant Biol. 36, 158–167. [DOI] [PubMed] [Google Scholar]
- Cooper, J.L. and Henikoff, S. (2004) Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 21, 1712–1718. [DOI] [PubMed] [Google Scholar]
- Cui, J. , Zhang, Z. , Shao, Y. , Zhang, K. , Leng, P. and Liang, Z. (2015) Genome‐wide identification, evolutionary, and expression analyses of histone H3 variants in plants. BioMed Res. Int. 2015, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draizen, E.J. , Shaytan, A.K. , Mariño‐Ramírez, L. , Talbert, P.B. , Landsman, D. and Panchenko, A.R. (2016) HistoneDB 2.0: a histone database with variants—an integrated resource to explore histones and their variants. Database, 2016 https://www.ncbi.nlm.nih.gov/research/HistoneDB2.0/index.fcgi/type/H3/variant/cenH3/#msa_div_browse [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell, J.M. (2010) Haploids in flowering plants: Origins and exploitation. Plant Biotechnol. J. 8, 377–424. [DOI] [PubMed] [Google Scholar]
- Dwivedi, S.L. , Britt, A.B. , Tripathi, L. , Sharma, S. , Upadhyaya, H.D. and Ortiz, R. (2015) Haploids: constraints and opportunities in plant breeding. Biotechnol. Adv. 33, 812–829. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W.C. and Rothfield, N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma, 91, 313–321. [DOI] [PubMed] [Google Scholar]
- Fachinetti, D. , Folco, H.D. , Nechemia‐Arbely, Y. , Valente, L.P. , Nguyen, K. , Wong, A.J. , Zhu, Q. et al (2013) A two‐step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg, G. and Mirny, L.A. (2012) Higher‐order chromatin structure: bridging physics and biology. Curr. Opin. Genet. Develop. 22, 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa, T. and Earnshaw, W.C. (2014) The centromere: chromatin foundation for the kinetochore machinery. Develop. Cell, 30, 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles, L.M. , Khaled, A. , Laffaire, J.B. , Chaignon, S. , Gendrot, G. , Laplaige, J. , Bergès, H. et al (2017) Loss of pollen‐specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 36, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. , Ahmad, K. , Platero, J.S. and van Steensel, B. (2000) Heterochromatic deposition of centromeric histone H3‐like proteins. Proc. Natl. Acad. Sci. USA, 97, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T. , Karimi‐Ashtiyani, R. and Houben, A. (2016) Haploidization via chromosome elimination: means and mechanisms. Annu. Rev. Plant Biol. 67, 421–438. [DOI] [PubMed] [Google Scholar]
- Karimi‐Ashtiyani, R. , Ishii, T. , Niessen, M. , Stein, N. , Heckmann, S. , Gurushidze, M. , Banaei‐Moghaddam, A.M. et al (2015) Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. USA, 112, 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A. , Mezulis, S. , Yates, C.M. , Wass, M.N. and Sternberg, M.J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Proto. 10, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher, T. , Starr, D. , Wang, W. , McCuiston, J. , Zhong, H. , Nuccio, M.L. and Martin, B. (2016) Maternal haploids are preferentially induced by CENH3‐tailswap transgenic complementation in maize. Front. Plant Sci. 7, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher, T. , Starr, D. , Richbourg, L. , Chintamanani, S. , Delzer, B. , Nuccio, M.L. , Green, J. et al (2017) MATRILINEAL, a sperm‐specific phospholipase, triggers maize haploid induction. Nature, 542, 105–109. [DOI] [PubMed] [Google Scholar]
- Khanday, I. , Skinner, D. , Yang, B. , Mercier, R. and Sundaresan, V. (2019) A male‐expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature, 565, 91. [DOI] [PubMed] [Google Scholar]
- Kuppu, S. , Tan, E.H. , Nguyen, H. , Rodgers, A. , Comai, L. , Chan, S.W.L. and Britt, A.B. (2015) Point mutations in centromeric histone induce post‐zygotic incompatibility and uniparental inheritance. PLoS Genet. 11, e1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova, I. , Schubert, V. , Fuchs, J. , Klatte, S. , Macas, J. and Schubert, I. (2006) Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell, 18, 2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova, I. , Kuhlmann, M. , Friedel, S. , Rutten, T. , Heckmann, S. , Sandmann, M. , Demidov, D. et al (2013) Arabidopsis KINETOCHORE NULL2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell, 25, 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova, I. , Sandmann, M. and Demidov, D. (2014) Centromeres and kinetochores of Brassicaceae. Chromosome Res. 22, 135–152. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Wang, Y. , Ren, J. , Mei, M. , Frei, U.K. , Trampe, B. and Lübberstedt, T. (2016) Maize doubled haploids. Plant Breed. Rev. 40, 123–166. [Google Scholar]
- Liu, C. , Li, X. , Meng, D. , Zhong, Y. , Chen, C. , Dong, X. , Xu, X. et al (2017) A 4‐bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant, 10, 520–522. [DOI] [PubMed] [Google Scholar]
- Logsdon, G.A. , Barrey, E.J. , Bassett, E.A. , DeNizio, J.E. , Guo, L.Y. , Panchenko, T. , Dawicki‐McKenna, J.M. et al (2015) Both tails and the centromere targeting domain of CENP‐A are required for centromere establishment. J. Cell Biol. 208, 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K. (2003) Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Develop. 13, 127–135. [DOI] [PubMed] [Google Scholar]
- Luger, K. , Mäder, A.W. , Richmond, R.K. , Sargent, D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251. [DOI] [PubMed] [Google Scholar]
- Maheshwari, S. , Tan, E.H. , West, A. , Franklin, F.C.H. , Comai, L. and Chan, S.W.L. (2015) Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 11, e1004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhis, N. , Jones, S.J. and Gsponer, J. (2019) Improved measures for evolutionary conservation that exploit taxonomy distances. Nat. Commun. 10, 1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec, M. and Hensel, G. (2018) Targeted base editing systems are available for plants. Trends Plant Sci. 23, 955–957. [DOI] [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi, M. and Chan, S.W.L. (2010) Haploid plants produced by centromere‐mediated genome elimination. Nature, 464, 615–618. [DOI] [PubMed] [Google Scholar]
- Ravi, M. , Kwong, P.N. , Menorca, R.M.G. , Valencia, J.T. , Ramahi, J.S. , Stewart, J.L. , Tran, R.K. et al (2010) The rapidly evolving centromere‐specific histone has stringent functional requirements in Arabidopsis thaliana . Genetics, 186, 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi, M. , Marimuthu, M.P.A. , Tan, E.H. , Maheshwari, S. , Henry, I.M. , Marin‐Rodriguez, B. , Urtecho, G. et al (2014) A haploid genetics toolbox for Arabidopsis thaliana. Nat. Commun. 5, 1‐16. [DOI] [PubMed] [Google Scholar]
- Ren, J. , Wu, P. , Trampe, B. , Tian, X. , Lubberstedt, T. and Chen, S. (2017) Novel technologies in doubled haploid line development. Plant Biotechnol. J. 15, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, M. , Kajala, K. , Pauluzzi, G. , Wang, D. , Reynoso, M.A. , Zumstein, K. , Garcha, J. et al (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type‐specific gene expression and function using tomato as a model. Plant Physiol. 166, 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanei, M. , Pickering, R. , Kumke, K. , Nasuda, S. and Houben, A. (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. USA, 108, E498–E505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui‐Simarro, J.M. , Corral‐Martinez, P. , Parra‐Vega, V. and Gonzalez‐Garcia, B. (2011) Androgenesis in recalcitrant solanaceous crops. Plant Cell Rep. 30, 765–778. [DOI] [PubMed] [Google Scholar]
- Stoler, S. , Keith, K.C. , Curnick, K.E. and Fitzgerald‐Hayes, M. (1995) A mutation in CSE4, an essential gene encoding a novel chromatin‐associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Develop. 9, 573–586. [DOI] [PubMed] [Google Scholar]
- Tachiwana, H. , Osakabe, A. , Kimura, H. and Kurumizaka, H. (2008) Nucleosome formation with the testis‐specific histone H3 variant, H3t, by human nucleosome assembly proteins in vitro. Nucleic Acids Res. 36, 2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana, H. , Kagawa, W. , Shiga, T. , Osakabe, A. , Miya, Y. , Saito, K. , Hayashi‐Takanaka, Y. et al (2011a) Crystal structure of the human centromeric nucleosome containing CENP‐A. Nature, 476, 232–235. [DOI] [PubMed] [Google Scholar]
- Tachiwana, H. , Osakabe, A. , Shiga, T. , Miya, Y. , Kimura, H. , Kagawa, W. and Kurumizaka, H. (2011b) Structures of human nucleosomes containing major histone H3 variants. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 578–583. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B. , Masuelli, R. , Tyagi, A.P. , Comai, L. and Henikoff, S. (2002a) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell, 14, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B. , Masuelli, R. , Tyagi, A.P. , Comai, L. and Henikoff, S. (2002b) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell, 14, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, E.H. , Henry, I.M. , Ravi, M. , Bradnam, K.R. , Mandakova, T. , Marimuthu, M.P. , Korf, I. et al (2015) Catastrophic chromosomal restructuring during genome elimination in plants. Elife, 4, e06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraev, A. , Forster, B.P. and Jain, S.M. (2009) Advances in Haploid Production in Higher Plants. Berlin/Heidelberg, Germany: Springer. [Google Scholar]
- Tsunaka, Y. , Kajimura, N. , Tate, S.‐I. and Morikawa, K. (2005) Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 3, 3424–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veken, J. , Eeckhaut, T. , Baert, J. , Ruttink, T. , Maudoux, O. , Werbrouck, S. and Van Huylenbroeck, J. (2019) Cichorium intybus L.× Cicerbita alpina Walbr.: doubled haploid chicory induction and CENH3 characterization. Euphytica, 215, 134. [Google Scholar]
- Wang, N. and Dawe, R.K. (2018) Centromere size and its relationship to haploid formation in plants. Mol. Plant, 11, 398–406. [DOI] [PubMed] [Google Scholar]
- Yao, L. , Zhang, Y. , Liu, C. , Liu, Y. , Wang, Y. , Liang, D. , Liu, J. et al (2018) OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants, 4, 530–533. [DOI] [PubMed] [Google Scholar]
- Zhong, Y. , Liu, C. , Qi, X. , Jiao, Y. , Wang, D. , Wang, Y. , Liu, Z. et al (2019) Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants, 5, 575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Map of CENH3 histone fold domain showing amino acid substitutions and their haploid inducing frequency.
Figure S2 Alignment of D. melanogaster, S. cerevisiae, Arabidopsis thaliana, and H. sapiens CENH3 homologs.
Figure S3 Predicted structure of Arabidopsis thaliana CenH3 showing the regions of 2 and 11 amino acid deletions.
Figure S4 Sequence alignment of the HFD of CENH3 from 53 different angiosperm species.
Figure S5 Illustrating the conservation across eukaryotes of the alphaN helix among conventional H3’s, but not CENH3 (Draizen et al., 2016).
Figure S6 Pooled data from immunolocalization analysis of wild‐type and variant CENH3 proteins on the nucleus isolated from 10 days old seedlings.
Table S1 Showing all the point mutants tested so far for their ability to complement endogenous CENH3 null, set seeds and induce haploid upon out crossing to Ler gl1 carrying wild‐type CENH3
Table S2 Assessing seed death upon selfing for non‐complementing mutation
Table S3 Haploid induction frequencies of double point mutants tested
Table S4 Percent seed death and haploid induction rate of crosses where the variant CENH3 complementing endogenous cenh3 null was used either as a female (a) or a male (b) parent
Methods S1 This is a diagram showing where we chose to mutate amino acids, rather than an experimental result it summarizes what is described in the paper


