Figure 1.
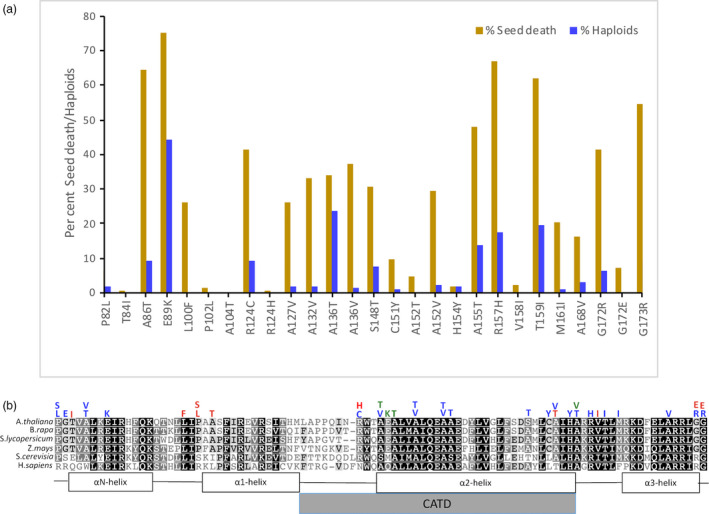
(a) Seed death and haploid induction upon outcrossing. Plants null for the endogenous cenh3 allele carrying a transgene with single amino acid substitutions in the CENH3 histone fold domain were pollinated by wild‐type CENH3 plants carrying the gl1 mutation. Seeds from the cross were assessed for seed death. Per cent haploid induction was calculated as the number of glabrous progeny among all live progeny. (b) Haploid induction rin plants carrying single amino acid substitutions in conserved residues of the CENH3 histone fold domain. Alignment of CENH3 HFD from three different dicot species and a monocot. S. cerevisiae and human CENH3 (CENP‐A) are included to highlight the level of conservation across kingdoms. The coloured letters above the alignment represent single amino acid substitutions tested at that position. Blue represents haploid inducers, red represents substitutions that complement the null allele but do not induce haploids, and green represents the single amino acid substitutions that fail to complement the null allele (lethal alleles). CATD is the CENP‐A (human CENH3) targeting domain. The following mutants were published earlier: P82S, G83E, A86V, P102S, A132T, A137T and G173E (Kuppu et al, 2015). Position of the αN helix, beginning with TVAL, is based on the human H3.1 nucleosome crystal structure.
