FIGURE 3.
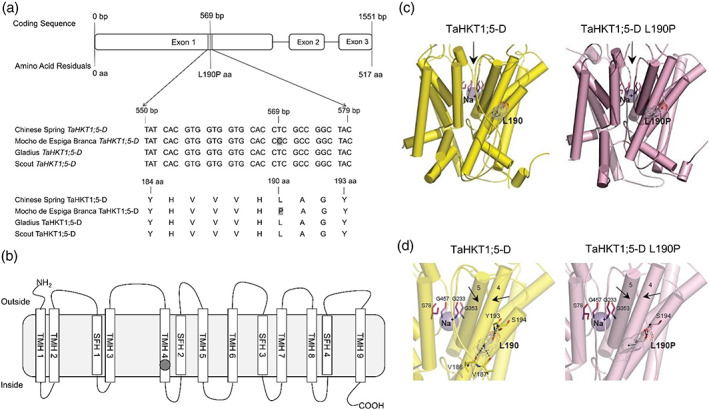
A SNP in Mocho de Espiga Branca TaHKT1;5‐D results in a L190P amino acid residue variation in the Na+ transporter TaHKT1;5‐D. (a) Partial alignment of TaHKT1;5‐D coding and amino acid sequences in Mocho de Espiga Branca, Gladius and Scout compared to Chinese Spring. (b) Schematic of the TaHKT1;5‐D protein showing the transmembrane α‐helices (TMH 1–9) and selectivity filter α‐helices (SFH 1–4) adapted from Xu et al. (2018); the Na+ selectivity filter motif S78‐G233‐G353‐G457 indicated in blank circles and location of the L190P variant indicated in a grey circle. (c) Molecular models of TaHKT1;5‐D (left, yellow) and TaHKT1;5‐D L190P (right, salmon) transport proteins in cartoon representations with cylindrical α‐helices illustrating 3D folds. Constrictions in selectivity filters are bound by four residues (cpk magenta sticks) that contain Na+ ions (violet spheres). Black arrows illustrate directional flows of Na+ that are likely to enter the permeation trajectory by‐passing selectivity filter constrictions. Variant residues L190 and L190P (cpk sticks and dots, bold types) between wild‐type TaHKT1;5‐D and the L190P mutant are indicated; the dots illustrate volumes of van der Waals radii. (d) Detailed views of α‐helices, which neighbour selectivity filter constriction, containing Na+ (violet spheres), located near selectivity filter residues S78, G233, G353, G457 (cpk magenta) for TaHKT1;5‐D (left) and the L190P mutant (right), which are crucial for permeation function. In each protein, polar contacts (cpk sticks and dots) of L190 (TaHKT1;5‐D) and L190P (TaHKT1;5‐D L190P) positioned on α‐helices 4, are indicated by dashed lines (separations between 2.6 Å and 3.2 Å) [Colour figure can be viewed at wileyonlinelibrary.com]
