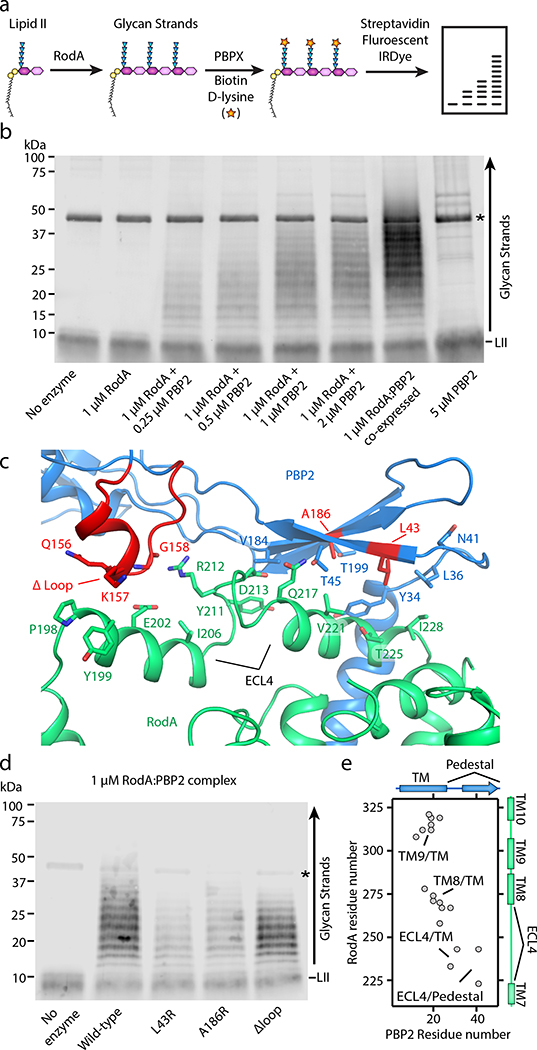
Figure 2 |. PBP2 activates RodA GT activity through its pedestal domain.
a, Graphical representation of polyacrylamide gel-based assay to detect RodA GT activity in vitro. The terminal residues in the stem peptide of the polymerized glycan strands and the lipid II percursor are exchanged with biotin-D-lysine via E. faecalis PBPX transpeptidase enzyme, transferred to a PVDF membrane, and then detected by fluorescently labeled streptadivin (Streptavidin-IRDye-800CW). b, RodA polymerizes lipid II only in the presence of PBP2. Asterisk represents the PBPX biotinylating enzyme. Note that complexes purified following co-expression are more active than when they are reconsitituted from individual preparations. Representative image of 1 of 3 experiments. c, Close up view of RodA-PBP2 extracytoplasmic interface II. Amino-acid substitutions are indicated in red. d, Substitutions in the PBP2 pedestal domain impair RodA GT activity in vitro. Representative image of 1 of 2 experiements. e, Evolutionary covariation map showing 19 evolutionary couplings between RodA and PBP2, generated from previously published data13.