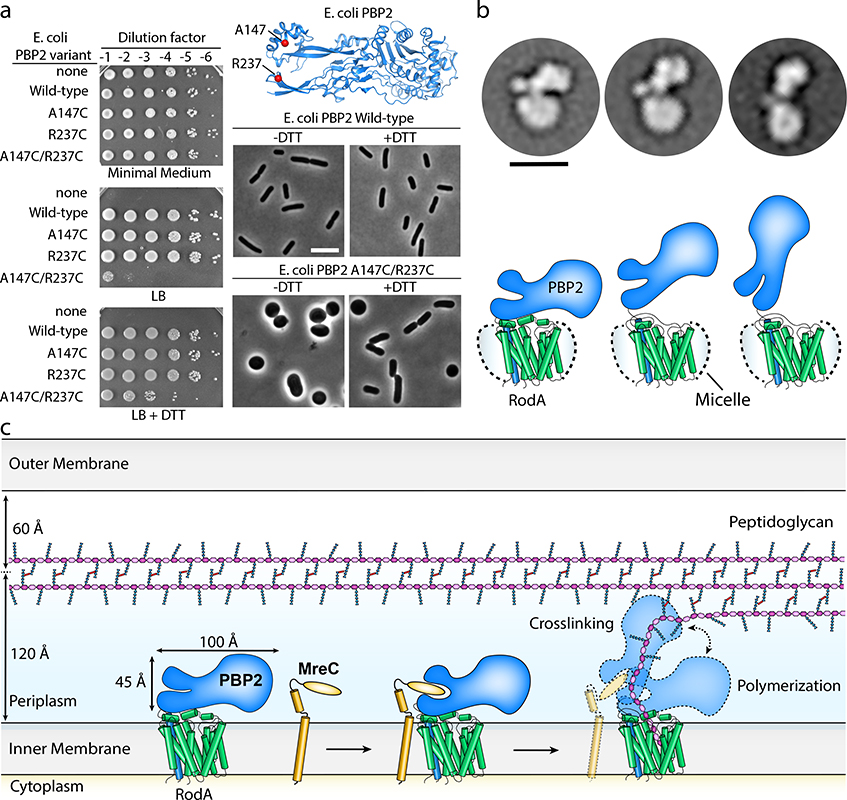
Figure 3 |. RodA:PBP2 complex adopts a broad range of conformations to facilitate cell wall synthesis.
a, Locking PBP2’s pedestal domain is lethal in E. coli. The top right panel shows the E. coli PBP2 structure15 (PDB code: 6G9P) highlighting the cysteine substitutions as red spheres. Overnight cultures of the E. coli strains in which PBP2-RodA had been deleted and complemented with a plasmid encoding RodA without PBP2 (“none”) or RodA with the PBP2 variants listed were serially diluted and spotted on either M9 Minimal Medium agar (Rod non-essential) or LB agar (Rod essential) in the presence or absence of 10 mM DTT (left panel). The bottom right panels show micrographs of E. coli cells inactivated for native PBP2 expressing either PBP2 (WT) or the cysteine variant from a plasmid. Scale bar, 5 μm. Results shown are derived from one experiment with n = 650 cells analyzed per group but are representative of two experiments. b, Representative negative-stain electron microscopy 2D class averages of RodA-PBP2 complex (top panel) depicting different conformations, which are schematically represented in the bottom panel. The images were collected on a Tecnai-T12 microscope and the resulting class averages were calculated from a total of 32,152 particles. Scale bar represents 10 nm. c, Model of RodA:PBP2-mediated peptidoglycan cell wall synthesis. The dimensions of the RodA:PBP2 complex structure are shown in relation to the Gram-negative bacterial cell envelope.