Fig. 4. The DNMT3B3-acidic patch interaction is required for chromatin recruitment.
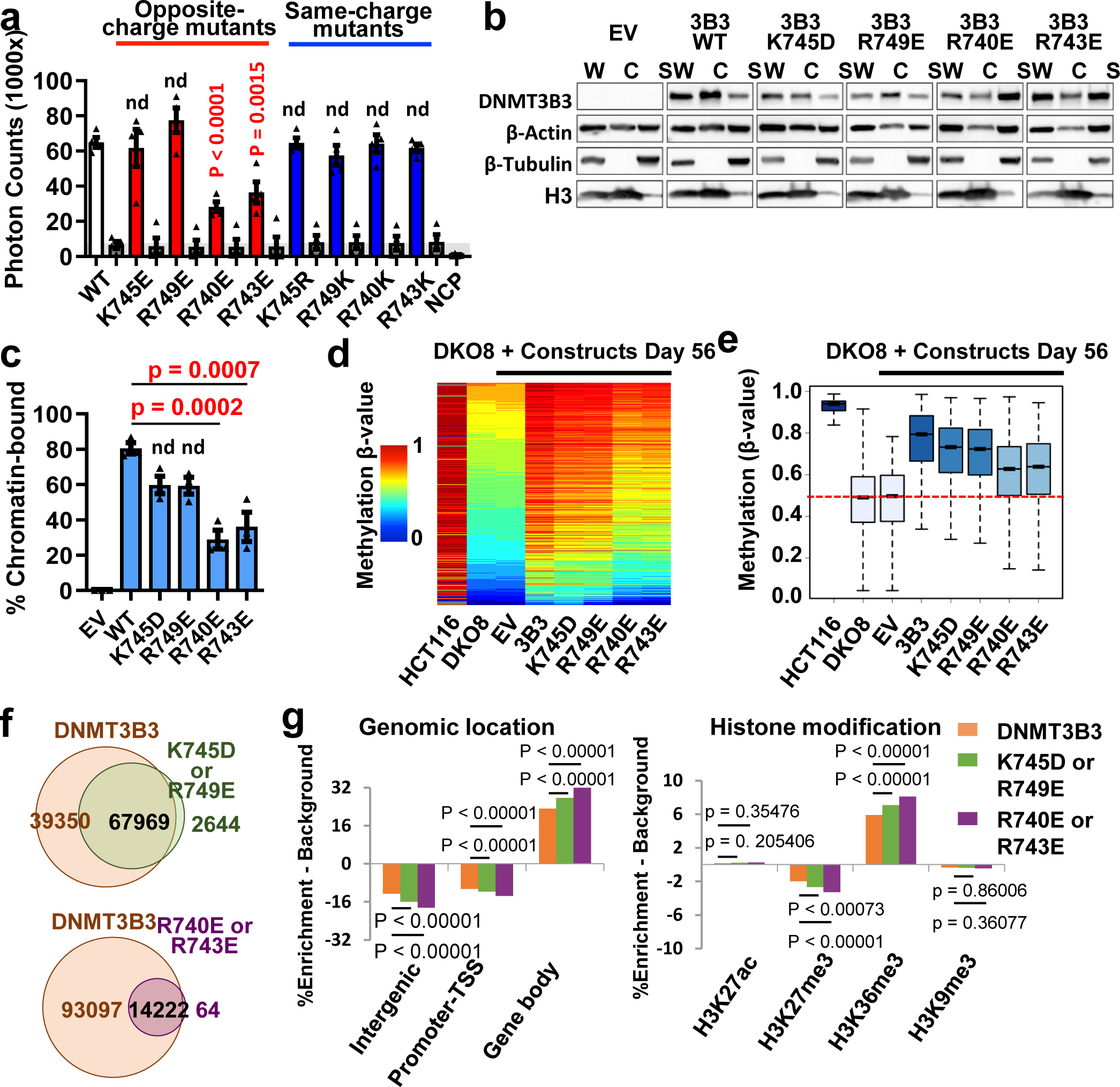
a, AlphaScreen interaction assay between His-tagged wildtype and CLD mutant DNMT3A2/DNMT3B3 complexes and biotinylated NCP. Opposite-charge mutants (red); same-charge mutants (blue); “DNMT only” and “NCP only” controls (grey). Data are mean ± sem, n = 4. b,c One representative western blot (b) and graphical representation from 3 independent biological replicates (c) of DNMT3 chromatin association. Chromatin was fractionated from DKO8 cells with introduced wildtype and mutant MYC-tagged DNMT3B3. The presence of DNMT3B3 in chromatin and soluble fractions was determined by immunoblotting. W: whole protein; C: chromatin fractions; S: soluble fractions. Data are mean ± sem, n = 3. d, Methylation heatmap of CpG sites in HCT116 wildtype and DKO8 cells expressing the indicated DNMT3B3 WT or CLD mutants. Methylation levels in HCT116 and DKO8 cells from two independent experiments are represented by color scale β-value (0–1, 0–100% methylated), where every row represents one CpG. e, Box plot of 109,998 probes, showing the distribution of DNA methylation levels for each indicated cell line. Median and interquartile range are represented by the bar and blue box; outliers outside the 5th to 95th percentiles are not shown. Dashed red line: median β-value in DKO8 cells. f, Venn diagrams showing total number of target methylation sites in DNMT3B3 wildtype and mutant cells. g, DNMT3B3 histone core-interaction mutations make methylation targeting more dependent on DNMT3-histone tail binding. Same 109,998 probes were used as in e. For DNMT3B3, n= 107319; for 745D/749E, n=70613; for 740E/743E, n=14286. Methylation becomes enriched at gene bodies, marked by PWWP-binding H3K36me3/me2 modifications, and decreased at transcription start sites (TSS), whose interactions are excluded by ADD interactions. p values: one-way ANOVA with Dunnett post-test (normality confirmed by Shapiro-Wilk test) (a and c) or two-sided z-test (g). For gel source data, see Supplementary Figure S2.
