FIGURE 5.
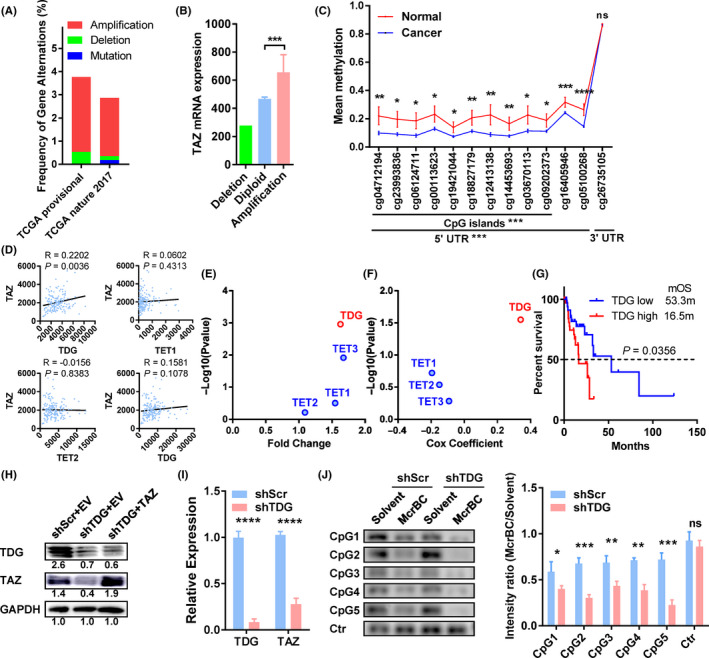
Thymine DNA glycosylase (TDG) regulates transcriptional coactivator with PDZ binding domain (TAZ) expression by DNA methylation of its CpG islands. A, Genetic changes in gastric cancer cohorts from The Cancer Genome Atlas (TCGA) provisional dataset (esophageal cancer [ESCA], n = 186) and TCGA nature 2017 dataset (esophageal/stomach cancer, n = 556). B, TAZ mRNA levels of esophageal cancer with different DNA copy numbers. C, Methylation rates of DNA around the TAZ gene in esophageal cancer and normal tissues. Probes and their locations are shown along the x‐axis. D, Correlation of TAZ mRNA (y‐axis) with demethylase mRNA (x‐axis) in esophageal cancer. E, Fold change of demethylases in esophageal tumor vs normal tissues (x‐axis), with P values indicated on the y‐axis. F, Association of demethylases with survival in esophageal cancer patients. G, Kaplan‐Meier curve of patient survival compared to their TDG expression. H, Protein levels of TDG and TAZ in the indicated cells. Results were quantified by ImageJ and shown below the bands. I, TDG and TAZ mRNA levels in TDG‐depleted and control cells. J, McrBC‐PCR results with five primers targeting the CpG islands of the TAZ gene in TDG‐depleted and control (Ctr) cells. Representative pictures are shown on the left, and quantification bars on the right. Data in A and B were extracted from cBioPortal. Data in C‐G were derived from TCGA ESCA methylation data, mRNA expression data, and clinical data. Data in H‐J resulted from in vitro studies. *P < .05, **P < .01, ***P < .001, ****P < .0001. EV, empty vector; ns, no significance; shScr, lentivirus containing scramble shRNA; shTDG, lentivirus containing shRNA against TDG; TAZ, plasmid containing TAZ
