Figure 3.
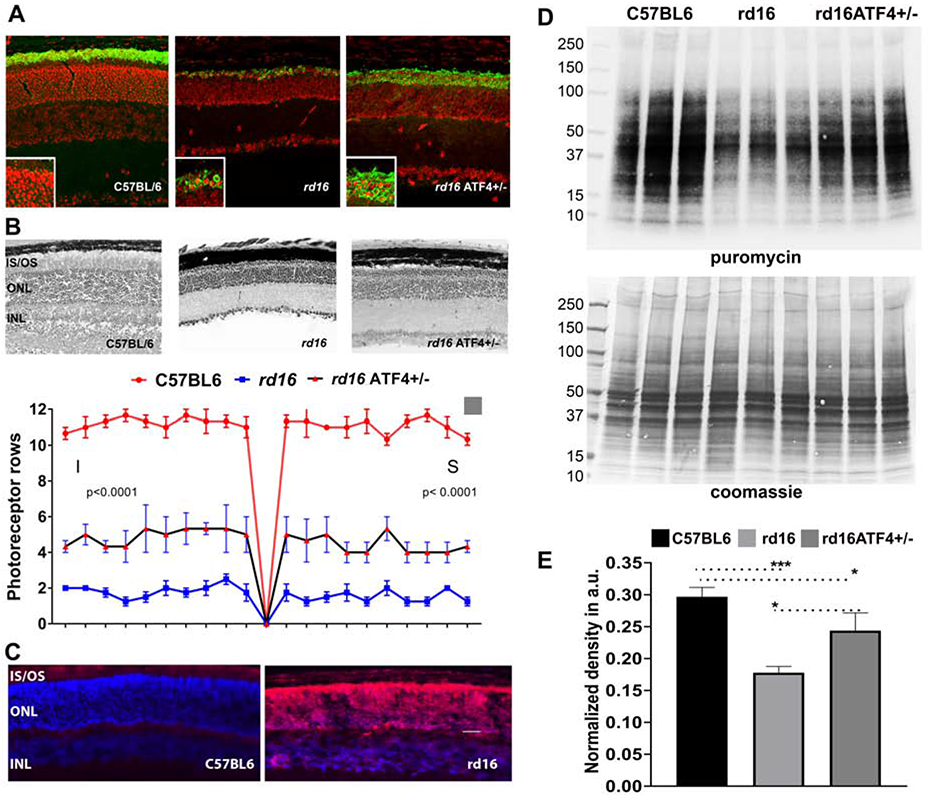
Molecular mechanisms of retinopathy in rd16 mice. Immunohistochemical analysis of C57BL6, rd16, and rd16 ATF4+/− retinas show mislocalization of rhodopsin in rd16 retinas at P18 (unpublished data). Detection of rhodopsin (green) was performed using primary anti-rhodopsin antibody ID4. Propidium iodide was used to stain retinal cell nuclei (red) (A). Delay of photoreceptor cell death in rd16 ATF4+/− retinas. H&E-stained retina sections are shown in the upper panel, and counts of photoreceptor rows are shown in the bottom panel (B). P15 rd16 retinas show significant accumulation of adenylyl cyclase (red) in the ONL. Based on the mechanism proposed by Nakao et al., this mislocalization could contribute to the retinal pathophysiology of rd16 mice (C). Study of the molecular mechanisms responsible for the delay of retinal degeneration in rd16 ATF4+/− retinas demonstrates enhanced synthesis of proteins in general (D) and rhodopsin in particular (A). Protein synthesis was assessed by the SUnSET method (Starr et al., 2019). C57BL6, rd16, and rd16 ATF4+/− mice were intraperitoneally injected with puromycin at a dosage of 0.04 μmol/g body mass. After 30 min of puromycin injection, retinas were harvested. Retinal proteins with incorporated puromycin were extracted in RIPA buffer, and their concentrations were estimated. Protein (60 μg) was separated by SDS-PAGE and transferred to a PVDF membrane, which was then probed with anti-puromycin antibody. A secondary antibody specific for IgG2a was also applied. As a loading control, membranes were stained with Coomassie blue after washing with distilled water (D). The relative densities of entire lanes were measured using ImageJ software. Puromycin densities were normalized to their corresponding Coomassie densities (E).
