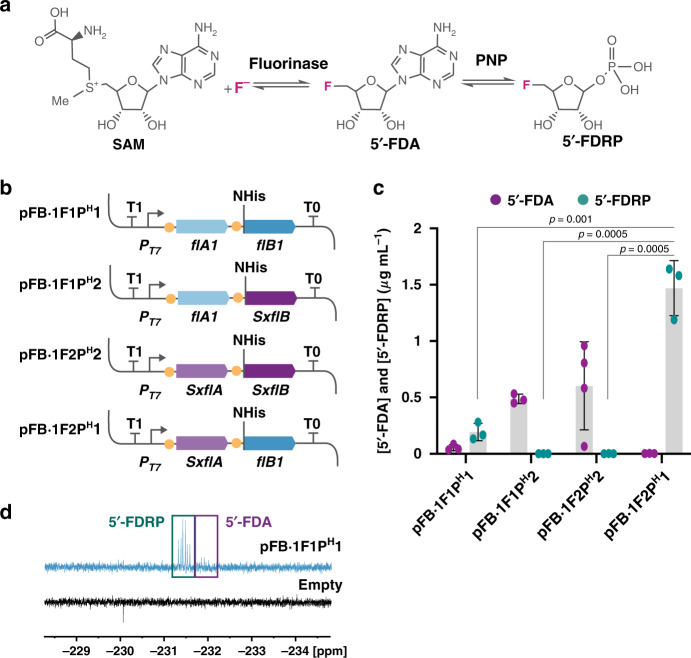
Fig. 4. In vitro biosynthesis of fluorosugars in cell-free extracts of engineered P. putida.
a Fluorination and phosphorylation steps in the canonical biofluorination pathway of S. cattleya. In the second step, 5′-fluoro-5′-deoxyadenosine (5′-FDA) is converted into 5′-fluoro-5′-deoxy-D-ribose 1-phosphate (5′-FDRP) by PNP (purine nucleoside phosphorylase). b Plasmid constructs of FluoroBricks encoding fluorinases and PNPs from Streptomyces sp. MA37 (flA1 and flB1) or S. xinghaiensis (SxflA and SxflB) under control of the PT7 promoter, which is used in combination with the synthetic FRS-T7 RNA polymerase circuit. Phosphorylases were added with a 6× histidine tag in the N terminus as indicated. c 5′-FDA and 5′-FDRP biosynthesis in cell-free extracts of P. putida KT2440::FRS-T7RNAP bearing the FluoroBrick constructs described in (b). Data are presented as mean values and error bars correspond to standard deviations of at least three different biological replicates; An unpaired t-test statistical comparison of the data identifies differences in 5′-FDRP biosynthesis between samples, p values are shown in the figure. d 19F-NMR spectrum of fluorometabolites in cell-free extracts of engineered P. putida expressing flA1 and flB1 using the synthetic circuit, compared to the spectrum of cells bearing the empty plasmid. Source data underlying c are provided as a Source Data file.