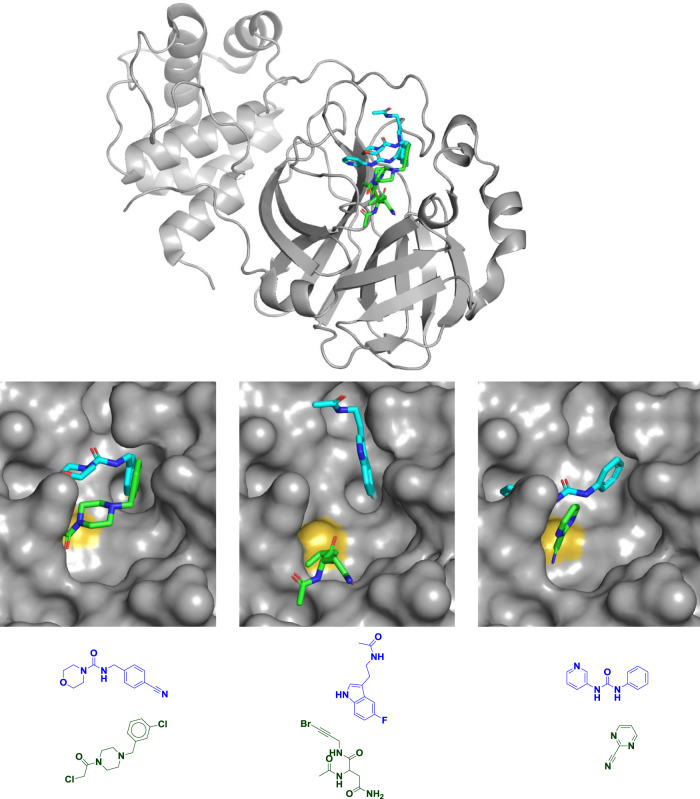
Fig. 1. Diverse fragments in diverse binding sites.
Mpro structure and chemical structures of six fragments bound to the active site of Mpro to illustrate their multiple binding locations. Although the catalytically active enzyme is an obligate homodimer, only one monomer is shown (top) for clarity. Covalent fragments are shown in green, noncovalent fragments are shown in blue or cyan, and the catalytic cysteine sulfur atom is shown in yellow. All six fragments are shown overlaid in the top panel, in pairs in the middle panel, and as chemical structures in the bottom panel; overlays are shown to illustrate the range of binding modes though each fragment was crystallized individually. All three noncovalent fragments shown come from a “poised” fragment library designed for rapid follow-up chemistry through the urea or amide linkages13. Most of the covalent fragment hits disclosed are chloroacetamides (bottom left), but others include bromoalkynes (bottom middle) and aromatic nitriles (bottom right). PDB codes are (from left to right and top to bottom): 5RFE, 5R7Z, 5R83, 5RET, 5RG3, and 5RHB. As noted by Douangamath et al.1, overlaying structures suggests many opportunities for fragment merging, for example the two compounds on the left.