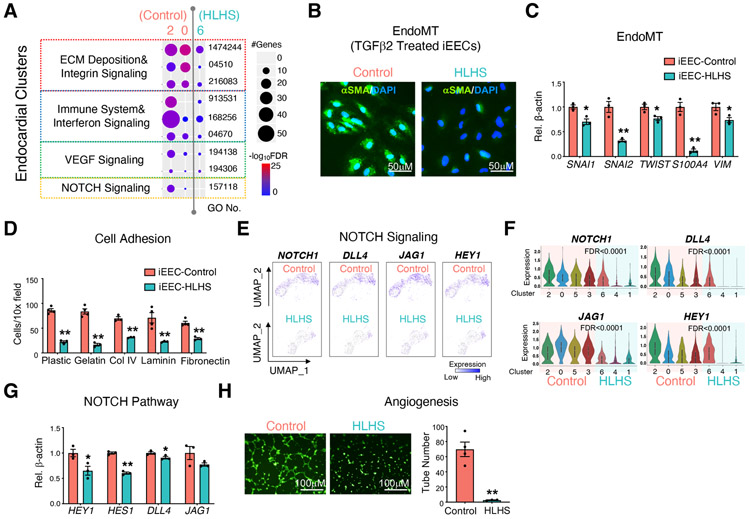
Figure 3. iPSC-ECs and iEECs revealed functional defects in the HLHS endocardium.
(A) Functional enrichment in endocardial clusters from control (cluster 2 & 0) and HLHS (cluster 6) iPSC-EC scRNA-seq. −log10FDR indicates the significance of enrichment. GO: gene ontology. α smooth muscle actin (αSMA) staining (B) and EndoMT related gene expression (C) in iEECs from control and HLHS patients after 7 days treatment with TGFβ2 (10 ng/μl). DAPI: nucleus. (D) Adhesion assay of iEECs with different ECMs to coat the culture dish. UMAP (E) and violin plots (F) of the represented genes of the NOTCH pathway from scRNA-seq. FDR indicated the significance of difference between control (cluster 2, 0, 5, and 3) vs HLHS (cluster 6, 4, and 1). (G) Expression levels of NOTCH pathway related genes were measured by qPCR in control and HLHS patient iEECs. (H) Tube formation of control and HLHS iEECs after seeding for 6 hr. iEECs were stained with Calcein AM (green) before seeding. Pictures are under 10X magnification. In (C), (D), (G), (H), data shown as the mean ± SEM. *p<0.05, **p<0.01, control (n=4) vs HLHS (n=3).