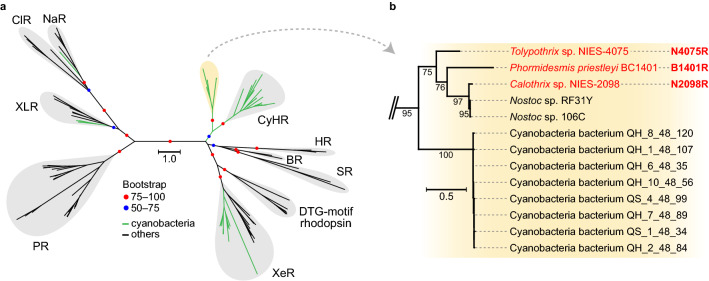
Figure 1.
The novel cyanorhodopsin clade. (a) Maximum likelihood tree of amino acid sequences of microbial rhodopsins. Bootstrap probabilities (≥ 50%) are indicated by colored circles. Green branches indicate cyanobacterial rhodopsins, and black branches indicate others. Rhodopsin clades are as follows: NaR (Na+-pumping rhodopsin), ClR (Cl−-pumping rhodopsin), XLR (xanthorhodopsin-like rhodopsin), PR (proteorhodopsin), XeR (xenorhodopsin), DTG-motif rhodopsin, SR (sensory rhodopsin-I and sensory rhodopsin-II), BR (bacteriorhodopsin), HR (halorhodopsin), CyHR (cyanobacterial halorhodopsin), and a novel cyanobacteria-specific clade (yellow shading). (b) Enlarged view of the novel cyanobacteria-specific clade. The three rhodopsins functionally examined in this study are shown in red. The scale bar represents substitutions per site.