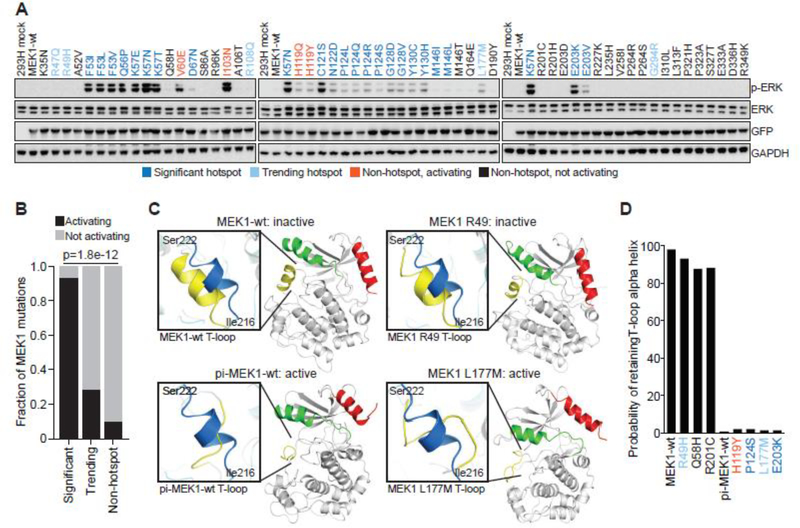
Figure 2. Concordance of biochemical and 3D modeling of MEK1 mutants.
A, GFP-tagged wildtype MEK1 (MEK1-wt) and MEK1 missense mutants were expressed in 293H cells. Expression of phosphorylated ERK (p-ERK), total ERK, GFP (MEK1) and GAPDH levels were assessed by western blot. Mutations were color coded as follows: statistically significant hotspot (dark blue); trending hotspot (light blue); non-hotspot, activating (orange); non-hotspot, not activating (black) B, Quantification of the accuracy of the computational inference of hotspot as compared to functional validation. Bars represent the fraction of either significant, trending, or not-significant mutants that were validated as activating in biochemical assays (black) versus not activating (gray). Statistical significance reflects enrichment of biochemical validation with hotspot prediction, p value = 1.8e−12. C, The structural changes to the MEK1 protein predicted to occur upon mutation of specific residues as assessed by molecular dynamics (MD) simulation. Results were compared to the simulated structure of MEK1-wt and phosphomimetic-MEK1-wt (pi-MEK1-wt). D, Quantitation of the probability that the natural T-loop alpha helix structure (inactive conformation) is retained following MD simulation of select MEK1 mutants.