Figure 4: The linc-SPRY3 family act as molecular sponges for IGF2BP3 preventing it from stabilizing HMGA2 and c-Myc.
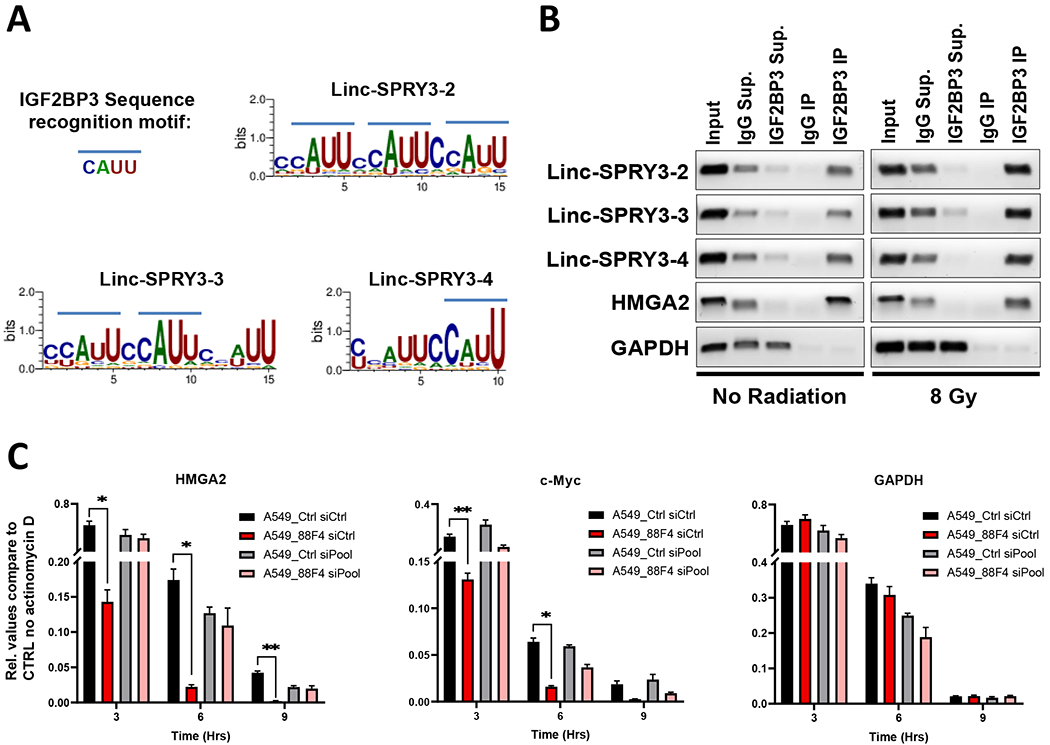
(A) Predicted binding motifs (using WebLogo3) of IGF2BP3 in each linc-SPRY3 transcript. (B) RT-PCR amplification of the recovered RNA from IGF2BP3 CLIP assays of untreated (No Radiation) or treated (8 Gy) H460 cells. The lanes are as follows (from left to right): Input RNA, IgG Supernatant, IGF2BP3 Supernatant, IgG Immunoprecipitation, IGF2BP3 Immunoprecipitation. HMGA is provided as a positive control and GAPDH as a negative control for IGF2BP3 interaction. Gel images are a representative of duplicate experiments. (C) Actinomycin D (ActD) RNA stability assays show rapid degradation of HMGA2 and c-Myc mRNA when the linc-SPRY3 RNAs are present (A549_88F4 siCtrl). siRNA knockdown rescues mRNA stability of HMGA2 and c-Myc (A549_88F4 siPool). GAPDH is provided as a negative control. Error bars represent SD from the mean (n=3)
