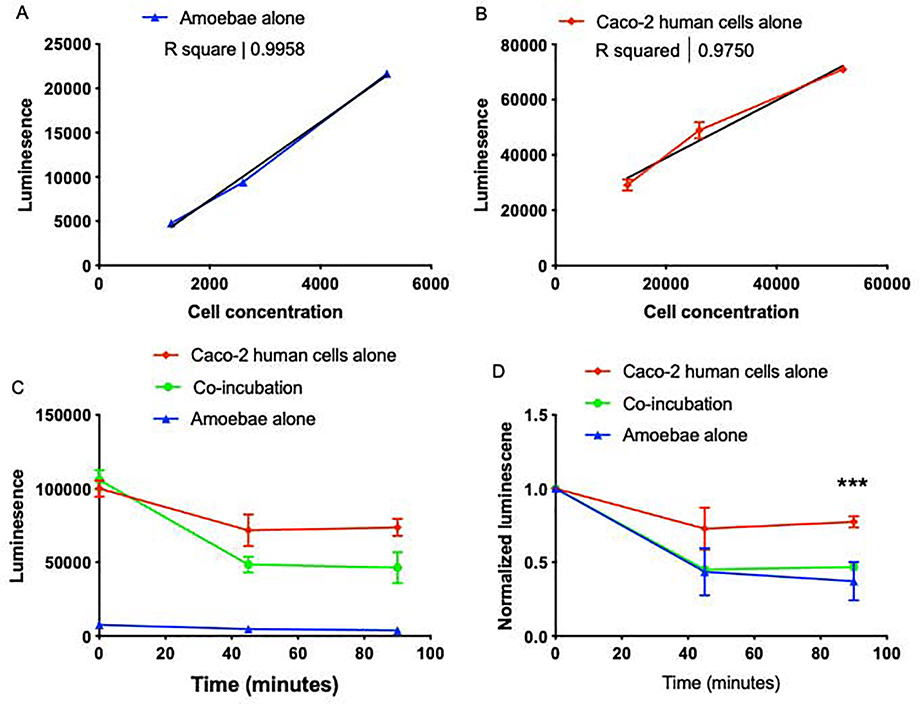
Figure 2: CellTiterGlo can be used to assay Caco-2 cell killing by amoebae.
Caco-2 cells were cultured and harvested as described [7]. 18–24 hours prior to performing the experiment, Caco-2 cells were plated in 96-well plates. On the day of the experiment, cells were washed with fresh TYI. Amoebae were added to create approximate co-incubation ratio of 1 amoeba: 10 Caco-2 cells. The CellTiterGlo assay (Promega) was carried out according to the manufacturer’s instructions. Three wells of amoebae alone, six wells of Caco-2 cells alone, and six wells of co-incubated samples were averaged to obtain one value per condition, and three experiments were performed independently on different days. (A) A dilution series of amoebae, or (B) human Caco-2 intestinal epithelial cells were assayed using CellTiterGlo. Best fit lines and R2 values are shown. CellTiterGlo signal correlates with the number of cells per well. Data represent the average values of two replicate wells for each cell concentration, and are representative of 3 independent experiments. (C) Amoebae were co-incubated (filled circles) with Caco-2 cells at an approximately 1:10 ratio, or amoebae (filled triangles) and Caco-2 cells (filled squares) were incubated separately as controls. Data represent the average values of two replicate wells for each sample, from one experiment. (D) Data from 3 independent experiments performed as in panel C were normalized to the value of each sample at Time = 0. There were statistically significant differences between the co-incubation and Caco-2 alone samples, as indicated. GraphPad Prism was for student’s unpaired t test statistical analysis. Mean values and standard deviations are shown; ns = P > 0.05, * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001.