Figure 3. Drivers and constraints in genome organization.
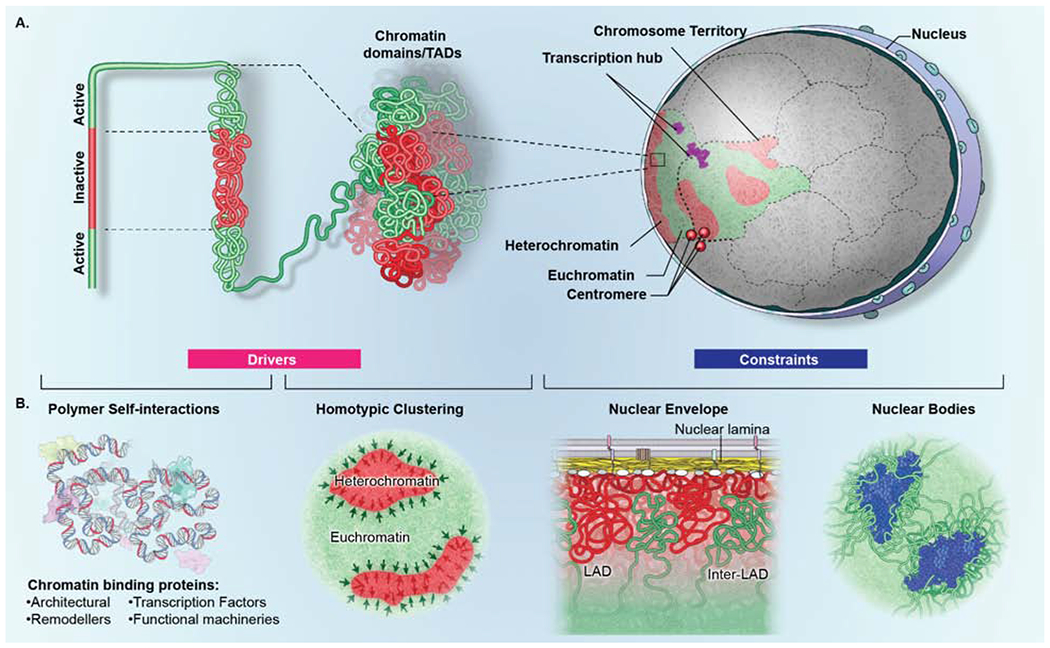
(A) Genome organization is determined by the interplay of drivers (pink box) and constraints (blue box). Higher-order organization is generated by self-interaction of transcriptionally active (green) and inactive (red) genome regions which fold onto themselves to form domains. Multiple homotypic (active or inactive) domains in turn aggregate both within a single chromosome and between domains on distinct chromosomes to form compartments. Homotypic interactions amongst multiple chromosomes generate transcription hubs (purple) and centromere aggregates (red spheres) which contain genes and centromeres located on distinct chromosomes, respectively. (B) (left) Drivers of genome organization include polymer-polymer interactions and phase separation. Homotypic polymer-polymer interactions among chromatin fibers form domains that are stabilized by phase separation (black arrows), including segregation of eu- and heterochromatin. (right) Constraints of genome organization include architectural features that physically interact with genome regions to influence their mobility and location, such as the nuclear lamina, which interacts with transcriptionally inactive lamina-associated domains (LADs) (red), interrupted by inter-LADs (green) or nuclear bodies, such as splicing speckles (blue), which preferentially associate with transcriptionally active chromatin.
