Figure 1. Plant immune responses.
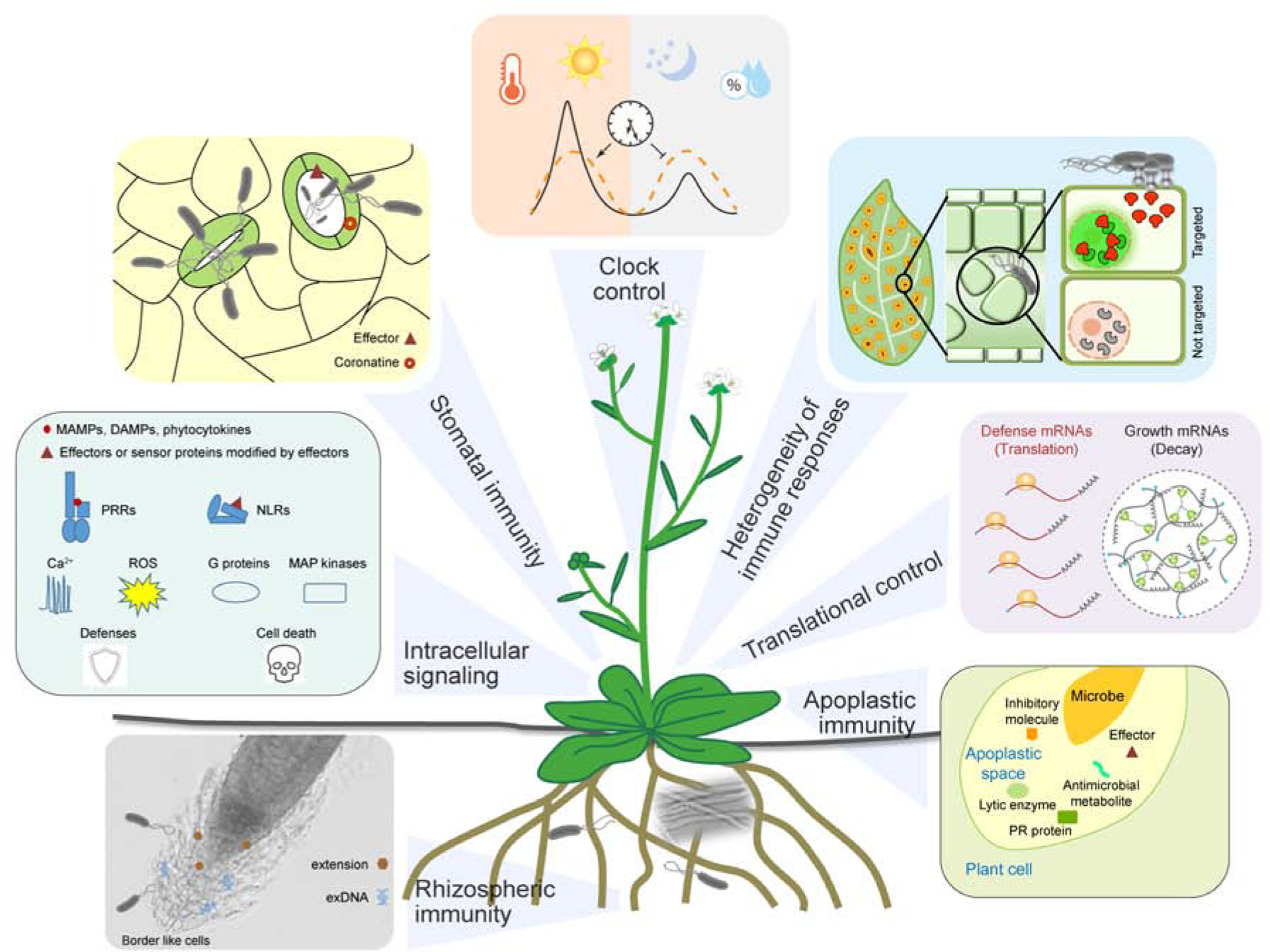
The front line of plant defense occurs in the stomata, rhizosphere and apoplast. Recognition of microbe-associated molecular patterns (MAMPs) can trigger stomatal closure to restrict entry of microbes into plants. To counteract, microbes produce effectors and coronatine to promote stomatal opening. In roots, extensins, border and border-like cells, and extracellular DNAs (exDNAs) contribute to rhizospheric immunity that functions in attracting beneficial microbes while repelling pathogens. In the apoplastic space, plants produce lytic enzymes to release immune-inducing MAMPs and antimicrobial pathogenesis-related (PR) proteins and metabolites to restrict the proliferation of invading pathogens. In the same space, microbes can use effectors and inhibitory molecules to counteract plants’ apoplastic immunity. Recognition of MAMPs and pathogen effectors activate pattern-recognition receptors (PRRs) on the cell surface and nucleotide-binding leucine-rich repeat immune receptors (NLRs) inside the cell to trigger pattern-triggered immunity (PTI) and effector-triggered immunity (ETI), respectively. These immune receptors signal through Ca2+, reactive oxygen species (ROS), G proteins, and MAP kinase cascades to confer resistance which in ETI is often associated with cell death. Activation of both PTI and ETI involve reprogramming of plant proteome through decay of house-keeping mRNAs (perhaps inside stress granules; circle) and activation of translation of defense proteins. Heterogeneity exists in plant responses to infection due to differential pathogen distribution and host immune sensitivity. Plant immune responses are also regulated by the circadian clock which not only helps plants anticipate infection, but also gates immune responses when infection occurs to minimize effects on plant physiology and fitness.
