Figure 1. Ca2+-activated potassium channels.
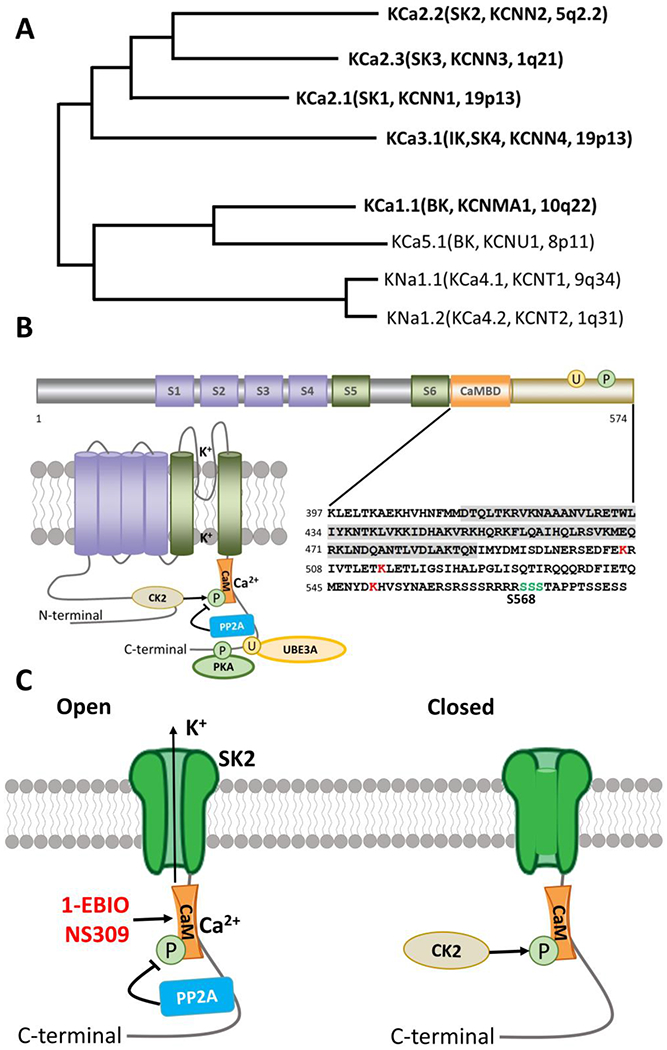
A. Phylogenetic tree for Ca2+ activated K channels. Modified from Aldrich R, Chandy KG, Grissmer S, Gutman GA, Kaczmarek LK, Wei AD, Wulff H. Calcium- and sodium-activated potassium channels, introduction. Last modified on 22/01/2015. Accessed on 16/08/2019. IUPHAR/BPS Guide to PHARMACOLOGY, http://www.guidetopharmacology.org/GRAC/FamilyIntroductionForward?familyld=69 (Aldrich, Chandy et al. 2019)
B. Functional domains, topography, and C-terminal sequence of SK2 channels. Cylinders represent α-helical transmembrane segments (S1-S6) with green (S5,S6) indicating the pore-lining segments. The C-terminal calmodulin binding domain (CaMBD) is shown in orange and shaded in detailed sequence (lower right panel). Major sites of PKA phosphorylation (P) and Ube3a ubiquitination sites (U) are indicated by green and red respectively in the detailed sequence (lower right panel). Phosphorylation and dephosphorylation of CaM by CK2 and PP2A are also indicated in the lower left panel.
C. Open and closed versions of the SK2 channel. PP2A and CK2 alter the Ca2+ sensitivity of the SK channels by dephosphorylating or phosphorylating SK-associated CaM. 1-EBIO and NS309 act by enhancing the Ca2+ sensitivity of SK channels.
