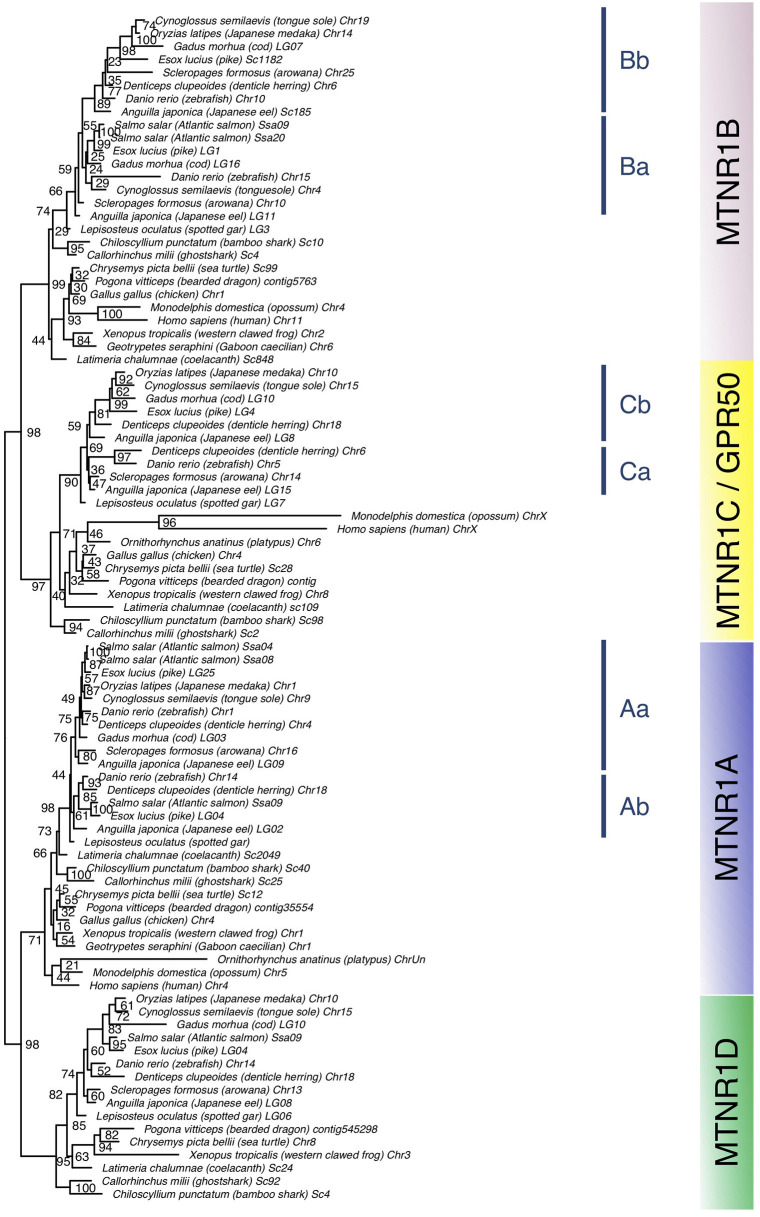
Figure 2.
Maximum-likelihood phylogeny tree of melatonin receptors of vertebrate representatives. Melatonin receptor phylogeny was inferred from alignment of the deduced amino-acid sequences of melatonin receptor using the PhyML algorithm with the AIC selection criteria of the Smart Model Selection and the tree Subtree Pruning and Regrafting (SPR) improvement algorithm. The four gnathostome monophyletic clades are indicated with different background colors. The blue line indicates teleost melatonin receptor clades. Branch nodes are supported by bootstrap analysis with 100 replicates and only nodes with bootstrap values above 50% were considered as supported.