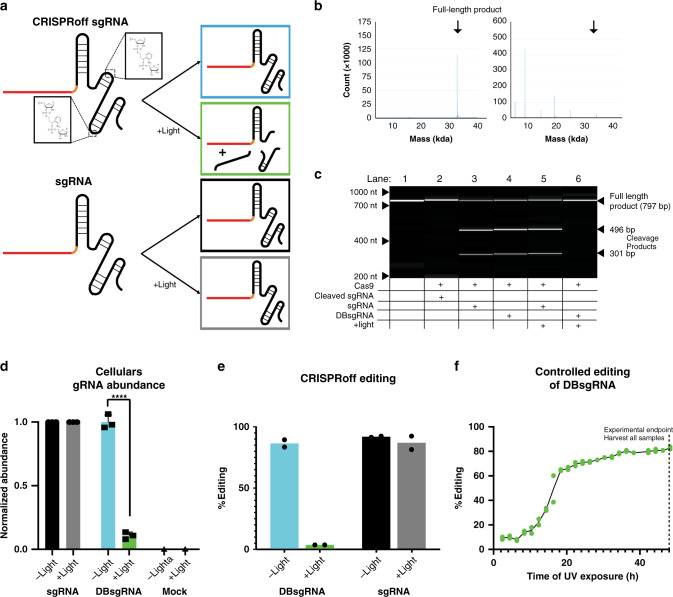
Fig. 1. Characterization of CRISPRoff sgRNAs.
a Schematic diagram of CRIPSRoff dual-breakage (DB) sgRNAs. Nucleotides at positions 57 and 74 are replaced with residues that are cleaved in response to light. In the dark DBsgRNAs (blue) perform like standard sgRNAs (black). Upon exposure to light, the DBsgRNAs fragment (green) and are no longer functional. b Electrospray ionization (ESI) analysis of sgRNAs. Left: DBsgRNA before exposure to light. Product is full-length DBsgRNA. Right: DBsgRNA after exposure to light. Amount of full-length DBsgRNA is significantly reduced and observed products match predicted weights based on cleavage locations. c DNA fragment analysis showing in vitro cleavage of DNA induced by RNPs formed with sgRNA or DBsgRNAs. When synthesized as individual parts (lane 2), DBsgRNAs are unable to support substrate cutting. Similarly, after exposure to light DBsgRNAs fail to support target cleavage (lane 6). In all cases, RNPs formed with standard sgRNAs are able to cleave substrate (lane 3, 5). d Amount of sgRNA in cells analyzed by ddPCR with or without light exposure. Level of standard sgRNAs did not decrease in response to light. In contrast, DBsgRNAs were significantly depleted from cells following irradiation. Mock transfections did not detect sgRNA in either condition, indicating primers were specific to the target. (n = 3 experimental replicates, data is presented as mean ±1 SD, ****p = 0.0003, Student’s two-tailed t-test with Bonferroni correction). e CRISPRoff editing in HEK293 cells at the DNMT1 locus, DBsgRNAs formed indels at rates similar to standard sgRNAs in dark conditions. When exposed to light, DBsgRNAs were no longer able to induce indels (n = 2 paired experimental replicates, data is presented as mean). f Editing time course of DBsgRNAs targeting DNMT1. Distinct cell samples were exposed to light once each, with 2-h intervals between samples and all cells were harvested at 48 h for genetic analysis (n = 2 technical replicates). CRISPRoff allowed for titratable levels of genome editing within populations. Source data are provided as a Source data file.