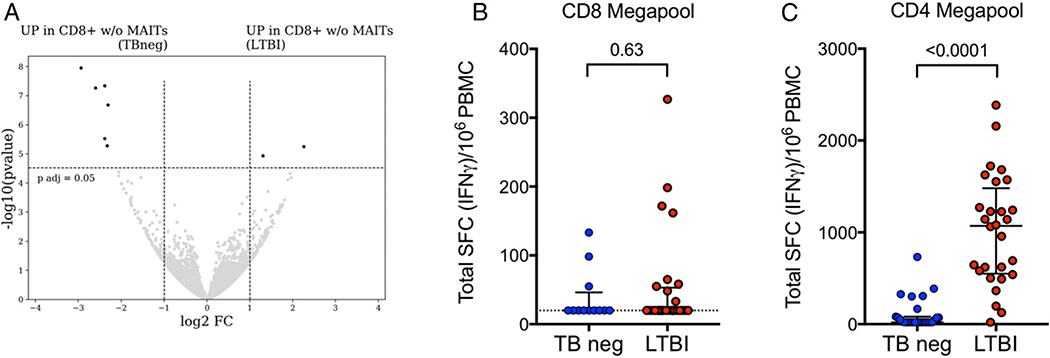
FIGURE 2. Minor differences between LTBI versus TB neg in M. tuberculosis–specific CD8+ T cell responses and transcriptomic analysis of memory CD8 T cells excluding Vα7.2+CD161+.
(A) Transcriptomic analysis of Vα7.2−CD161− memory CD8 T cells. Volcano plot obtained from the DEseq2 analysis showing log2 fold change versus −log10 p value. The eight differentially expressed genes are represented in black (adjusted p value <0.05 and absolute log2 fold change >1 are indicated by dotted lines). Genes upregulated in TB neg controls (left) and genes upregulated in LTBI (right). (B and C) Magnitude of epitope pool responses measured as total spot-forming cells (SCFs) per 106 PBMCs in an IFN-γ ELISPOT assay in individuals with LTBI and TB neg controls. Each dot represents one subject. Median ± interquartile range is indicated. Two-tailed Mann–Whitney U test. (B) CD8 megapool (LTBI, n = 28, red dots; TB neg, n = 12, blue dots), (C) CD4 megapool (LTBI, n = 28; TB neg, n = 27).