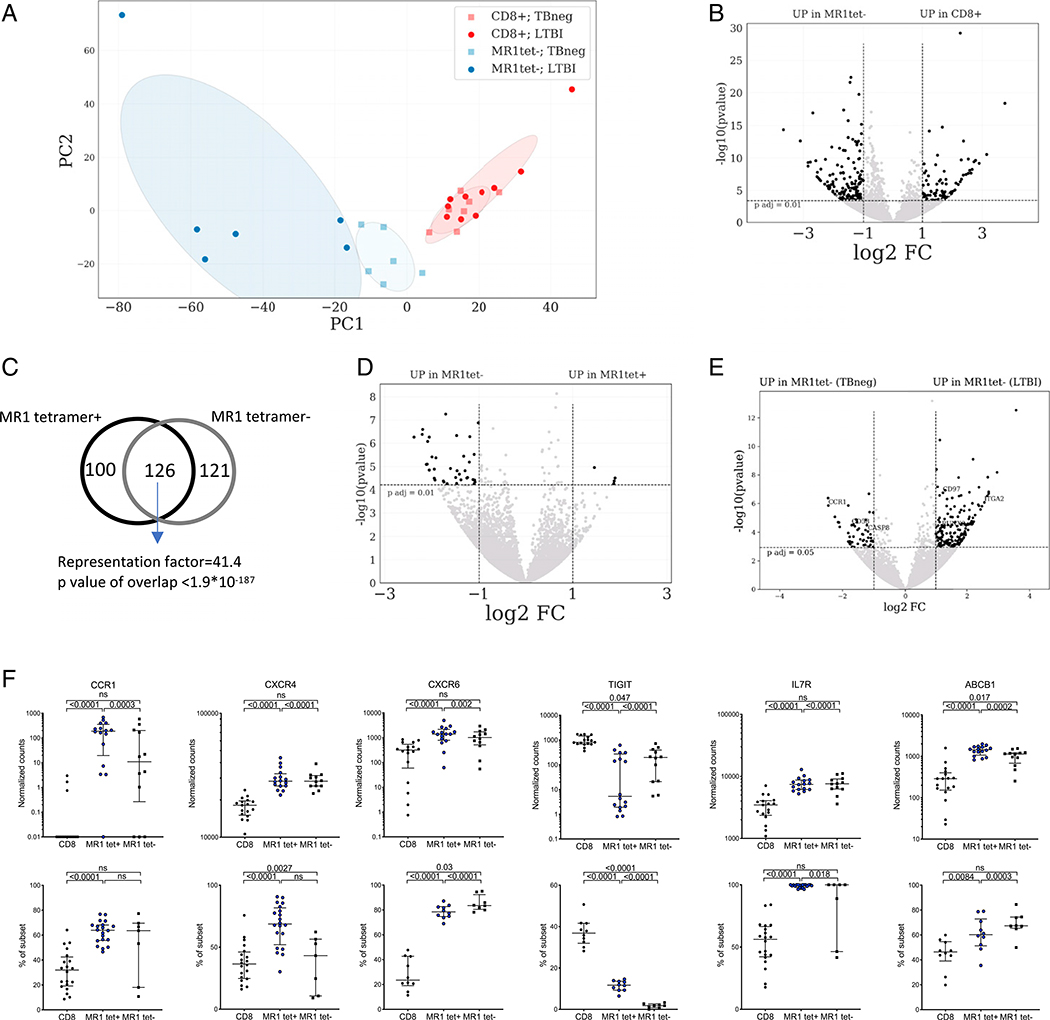
FIGURE 5. Gene expression profile and M. tuberculosis–specific signature of MR1tet− MAITs compared with memory CD8 T cells and MR1tet+ MAITs.
(A) PCA plot illustrating differences between memory CD8 T cells and MR1tet− MAITs and between LTBI and TB neg individuals. (B, D, and E) Volcano plots obtained from the DEseq2 analysis showing log2 fold change versus −log10 p value. The differentially expressed genes are represented in black [adjusted p value <0.01 (B and E) and p < 0.05 (D), absolute log2 fold change >1 are indicated by dotted lines]. (B) MR1tet− cells compared with memory CD8 T cells. (C) Venn-diagram showing overlap between the 226-gene signature identified in Fig. 4B and the signature in Fig. 5B, based on hypergeometric distribution test (considering the 18,315 transcripts detected within memory CD8 T cells as the total number of genes). (D) MR1tet− cells comparing individuals with LTBI versus TB neg. (E) Volcano plot comparing MR1tet− cells with MR1tet+ cells. (F) CCR1, CXCR4, CXCR6, TIGIT, IL-7R, and ABCB1 expression at the mRNA (upper panels: gene expression values in counts normalized by sequencing depth calculated by the DEseq2 package) and protein (lower panels: protein expression as percent frequency of subset) levels in memory CD8 T cells and MR1tet− MAITs. Gene expression data were derived from memory CD8 T cells from 17 individuals and MR1tet− cells (n individuals = 12) using an Illumina sequencing platform. Protein expression data were derived from memory CD8 T cells from 20 individuals and MR1tet− cells (n individuals = 7) using flow cytometry. Median ± interquartile range is shown. Two-tailed Mann–Whitney U test.