Abstract
Background:
Non-coding RNA-activated by DNA damage (NORAD), a novel identified lncRNA, was found to be aberrantly expressed in various types of cancer. This meta-analysis was performed to evaluate the value of lncRNA NORAD as a prognostic biomarker in human cancers.
Methods:
We searched PubMed, Web of Science, PMC, and Embase databases thoroughly for eligible literatures. Studies which explored the relationship of lncRNA NORAD expression with clinical outcomes in human cancers were included in our meta-analysis. Review Manager version 5.3 and Stata SE 12.0 were used to perform the data analyses.
Results:
Our meta-analysis results indicated that cancer patients with high lncRNA NORAD expression tended to have unfavorable overall survival (OS) (HR = 1.67; 95% CI, 1.44-1.95; P < 0.00001). Moreover, elevated lncRNA NORAD expression showed a significant relationship with poor tumor grade (OR = 1.61; 95% CI, 1.01-2.56; P = 0.05) and more lymph node metastasis (LNM) (OR = 2.66; 95% CI, 1.60-4.43; P = 0.0002).
Conclusions:
LncRNA NORAD could serve as a valuable biomarker to predict poor prognosis and LNM in various human tumors.
Keywords: LncRNA, NORAD, prognosis, cancer, meta-analysis
Introduction
Malignant cancers have been considered as a critical public health problem worldwide, resulting in a serious financial burden to society and families with cancer patients.1 It was reported that the estimated annual new cancer cases was 4.3 million in China, and the number of cancer deaths in China probably reached 2.8 million every year.2 Moreover, cancer patients with lymph node metastasis (LNM) and/or distant metastasis (DM) usually had worse 5-year survival. Despite the rapid progress in the management and treatment of cancers, the survival rate of cancer patients remains low.3-6 Therefore, it is a top priority to identify novel valuable biomarkers as predictors of cancer prognosis for cancer research.
With the development of sequencing and microarray technology, noncoding RNAs (ncRNAs) have been identified to be involved in the occurrence and development of human cancers.7-11 Long noncoding RNAs (lncRNAs) is a significant type of ncRNAs, commonly defined as RNA transcripts that contain more than 200 nucleotides but lack protein-coding potential.12 Numerous studies supported that lncRNAs were important players in the development and progression of various types of cancer.13,14 However, the lncRNAs which have been studied in-depth were relatively few until now; the function of most lncRNAs remains unclear.
Non-coding RNA-activated by DNA damage (NORAD, also named as linc00657) is a novel lncRNA, which was reported to be involved in regulation of the response to DNA damage, and the maintenance of genomic stability.15,16 In recent years, several studies have demonstrated that elevated lncRNA NORAD expression could promote the clinical progression and metastasis in multiple cancers17-19; but the combined prognostic value of lncRNA NORAD in cancer patients is still unclear. Therefore, we performed a comprehensive meta-analysis of eligible published studies to explore the association between lncRNA NORAD expression and clinical outcomes of solid tumors.
Materials and Methods
Search Strategy and Literature Selection
We conducted this meta-analysis followed the Preferred Reporting Items For Systematic Reviews and Meta-Analysis (PRISMA) statement20 (S1 Table) and AMSTAR (Assessing the methodological quality of systematic reviews) guidelines (S2 Table). Electronic searches of Pubmed, Web of Science, PMC, and Embase databases were performed to identify all articles that reported the relationship between lncRNA NORAD expression and prognosis in human cancers up to July 20, 2019. “LncRNA NORAD” OR “non-coding RNA-activated by DNA damage” OR “linc00657”, “cancer” OR “carcinoma”, and “prognosis” OR “survival” were used as keywords. The reference lists of eligible studies were also carefully scanned for potential supplements. The protocol was registered in the International Prospective Register of Systematic Reviews database (PROSPERO: CRD42019132009).
Eligibility and Exclusion Criteria
Two reviewers independently evaluated all eligible studies. Eligibility criteria for included studies were as follows: (i) evaluate the association of lncRNA NORAD expression with clinicopathological parameters and prognosis in patients with any type of cancer; (ii) provide sufficient data to obtain hazard ratios (HRs) with corresponding 95% confidence intervals (CIs) of overall survival (OS), or to calculate odds ratios (ORs) for clinicopathological parameters. Exclusion criteria were as follows: (i) non-human studies, non-English papers, reviews, letters, and case reports; (ii) studies without sufficient data for analysis; (iii) laboratory researches.
Data Extraction and Quality Assessment
Studies that met the inclusion criteria were evaluated for methodological quality by 2 independent reviewers following the Newcastle-Ottawa scale (NOS) scoring system,21 and then extracted for usable information. Disagreements were resolved through negotiation. The following information was recorded: last name of first author, year of publication, country, tumor type, sample size, specimens type, detection method, cutoff value, number of high/low lncRNA NORAD expression group, clinicopathological parameters, and HRs with corresponding 95% CIs for OS. HRs from multivariable analysis had priority to be considered; whereas studies only provided Kaplan-Meier survival curves, Engauge Digitizer version 4.1 was applied to extract usable data for calculation of the HRs and corresponding 95% CIs for OS using the method introduced by Tierney et al.22
Statistical Analysis
This meta-analysis was performed by use of Review Manager version 5.3 and Stata SE12.0 statistical softwares. The heterogeneity among the included studies was evaluated by I2 test and χ2-based Q test. HRs and corresponding 95% CIs were used to analyze the correlation between lncRNA NORAD expression and OS. ORs and corresponding 95% CIs were used to assess the relationship between lncRNA NORAD expression and clinicopathological parameters, such as gender, age, tumor size, tumor grade, tumor stage, and LNM. When pooled HRs or ORs were calculated, a fixed-effect model was used in case that no significant heterogeneity existed among studies (I2 < 50% and P > 0.05), otherwise a random-effect model was applied for the analysis. Probable publication bias was evaluated by Begg’s test, and sensitivity analysis was conducted by sequentially removing each study to confirm the stability of results. Statistical significance was defined as P < 0.05.
Results
Characteristics of Eligible Studies
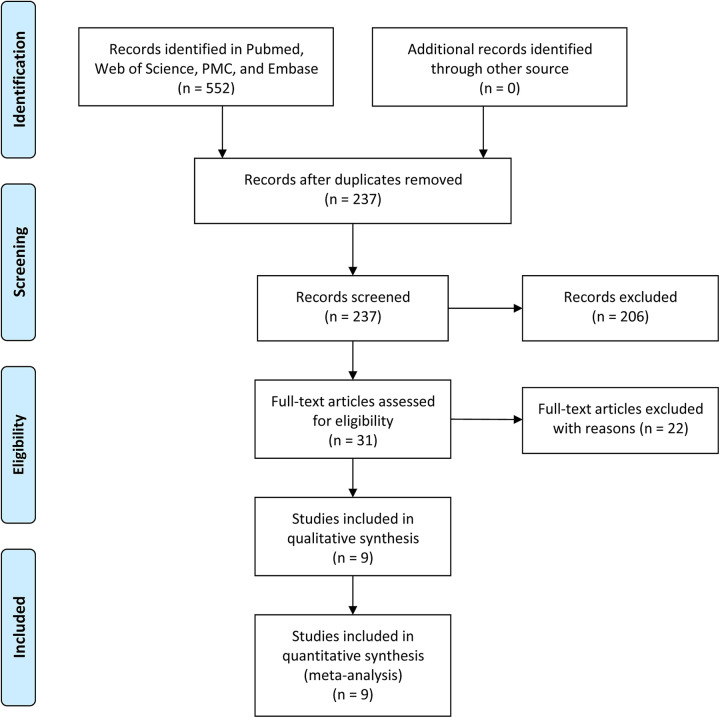
The flow chart diagram of literature search process is shown in Figure 1. A total of 552 reports were firstly identified in the electronic search of the above-mentioned databases. Following, 237 reports were retained by removing duplicates. After screening titles and abstracts, 206 papers were directly excluded. Additional 22 reports were excluded for not original studies or insufficient data. Finally, 9 available reports were included in our meta-analysis. The basic characteristics of these included studies were summarized in Table 1.
Figure 1.
Flow diagram showed the selection process of included studies.
Table 1.
Basic Characteristics of the Included Studies.
| Study/year | Country | Tumor type | Sample size | Specimens | Method | Cutoff value | NORAD expression | Age (high/low) | Male (high/low) Female (high/low) | Tumor size (high/low) | Tumor grade (high/low) | Tumor stage (high/low) | LNM | Outcome | HR | NOS | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | High | Low | |||||||||||||||
| Hu, 2017 | China | HCC | 49 | Tissue | qRT-PCR | Median | 24 | 25 | < 60y, 11/11; ≧ 60y, 13/14 | 19/18; 5/7 |
< 5cm, 15/4; ≥ 5cm, 9/21 |
NR | TNM stage: I-II, 17/6; III-IV, 7/19 | NR | NR | OS | SC | 7 |
| Huo, 2018 | China | Cervical cancer | 47 | Tissue | qRT-PCR | Median | 24 | 23 | < 50y, 9/11; ≧ 50y, 15/12 | NR | < 4cm, 8/14; ≥ 4cm, 16/9 | Well/Moderate, 15/19; Poor, 9/4 | FIGO stage: Ib-IIa, 8/17; IIb-IIIa, 16/6 | 12 | 3 | OS | SC | 7 |
| Lei, 2018 | China | Colon cancer | 80 | Tissue | qRT-PCR | Median | 40 | 40 | ≤ 55y, 15/22; > 55y, 25/18 | 23/27; 17/13 | < 5cm, 24/11; ≥ 5cm, 16/29 | NR | TNM stage: I-II, 30/18; III-IV, 10/22 | NR | NR | NR | NR | 6 |
| Li, 2017 | China | PC | 33 | Tissue | qRT-PCR | Median | 16 | 17 | < 60y, 8/7; ≧ 60y, 8/10 | 12/12; 4/5 |
< 2.5cm, 5/5; ≥ 2.5cm, 11/12 | NR | TNM stage: I-II, 14/15; III-IV, 2/2 | 6 | 8 | OS | SC | 7 |
| Li, 2018 | China | BC | 90 | Tissue | qRT-PCR | Mean | 37 | 53 | < 60y, 9/16; ≧ 60y, 28/37 | 31/37; 6/16 |
< 3cm, 24/39; ≥ 3cm, 13/14 | Low, 16/35; High, 21/18 |
Ta-T1, 14/33; T2-T4, 23/20 |
7 | 3 | OS | MA | 7 |
| Miao, 2018 | China | GC | 50 | Tissue | qRT-PCR | NR | 34 | 16 | < 45y, 11/5; ≧ 45y, 23/11 | NR | < 4cm, 19/9; ≥ 4cm, 15/7 | NR | NR | 13 | 2 | OS | SC | 7 |
| Wu, 2017 | China | ESCC | 106 | Tissue | qRT-PCR | Median | 53 | 53 | < 60y, 32/25; ≧ 60y, 21/28 | 42/40; 11/13 |
< 4cm, 30/42; ≥ 4cm, 23/11 |
Well/Moderate, 38/43; Poor, 17/10 |
UICC stage: I-II, 27/38; III, 26/15 |
27 | 18 | OS | MA | 8 |
| Yang, 2018 | China | HCC | 95 | Tissue | qRT-PCR | NR | 58 | 37 | < 60y, 37/17; ≧ 60y, 21/20 | 51/29; 7/8 |
< 5cm, 25/28; ≥ 5cm, 33/9 | I-II, 37/27; III-IV, 21/10 |
TNM stage: I, 38/28; II-III, 20/9 |
NR | NR | OS | MA | 8 |
| Zhang, 2018 | China | CRC | 47 | Tissue | qRT-PCR | Median | 24 | 23 | NR | NR | NR | NR | NR | NR | NR | OS | SC | 6 |
LNM: lymph node metastasis; HR: hazard ratios; NOS: Newcastle-Ottawa scale; PC: pancreatic cancer; BC: bladder cancer; GC: gastric cancer; ESCC: esophageal squamous cell carcinoma; HCC: hepatocellular carcinoma; CRC: colorectal cancer; qRT-PCR: quantitative real-time polymerase chain reaction; NR: not reported; OS: overall survival; SC: survival curve; MA: multivariate analysis.
All studies were published in 2017 or 2018, and conducted in China. The sample size ranged from 33 to 106 and a total of 597 cancer patients were included. Eight different types of cancer were estimated in the 9 eligible studies, including cervical cancer,23 colon cancer,24 pancreatic cancer (PC),18 bladder cancer (BC),19 gastric cancer (GC),25 esophageal squamous cell carcinoma (ESCC),26 hepatocellular carcinoma (HCC),27,28 and colorectal cancer (CRC).17 All the specimens were from tumor tissues and the lncRNA NORAD expression level was detected by qRT-PCR. Three studies directly reported the HRs with their 95% CIs of OS, and 5 studies provided survival curves that were used to calculate the HRs with their 95% CIs. All eligible studies were considered as high quality (NOS ≥ 6). And the included patients in each study were divided into high or low lncRNA NORAD expression group.
Association Between LncRNA NORAD Expression and OS
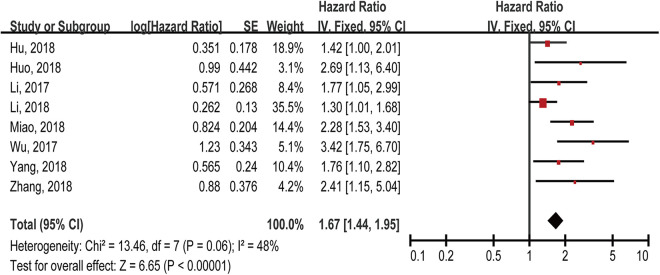
Eight studies directly or indirectly reported the HRs with their 95% CIs of OS for 517 cancer patients based on lncRNA NORAD expression levels. To explore the correlation between lncRNA NORAD expression and prognosis, a fixed-effect model was applied to calculate the combined HRs and corresponding 95% CIs because of no strong heterogeneity among these studies (I2 = 48%, P = 0.06). The pooled results showed that cancer patients with high lncRNA NORAD expression might be related to worse OS outcome in human cancers (HR = 1.67; 95% CI, 1.44-1.95; P < 0.00001) (Figure 2). A subgroup analysis of OS based on HR availability was also conducted and similar results were detected (Table 2). In addition, all the results indicated that higher lncRNA NORAD expression level might predict shorter survival for cancer patients.
Figure 2.
Forest plot for the relationship between lncRNA NORAD expression and OS.
Table 2.
Results of Association Between High NORAD Expression and Characteristics of Cancer Patients.
| Stratified analysis | No. of studies | No. of patients | Test of association | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|
| Pooled HR/OR 95% CI | p-value | I2 (%) | p-value | Model | |||
| OS | 8 | 517 | 1.67 (1.44, 1.95) | < 0.00001 | 48 | 0.06 | Fixed |
| Subgroup analysis of OS on HR availability | |||||||
| MA | 3 | 291 | 1.83 (1.12, 3.02) | 0.02 | 73 | 0.02 | Random |
| SC | 5 | 226 | 1.85 (1.49, 2.29) | < 0.00001 | 11 | 0.34 | Fixed |
| Age (young vs ≥ old) | 8 | 550 | 0.93 (0.63, 1.36) | 0.71 | 14 | 0.32 | Fixed |
| Gender (male vs. female) | 6 | 453 | 1.31 (0.84, 2.02) | 0.23 | 0 | 0.57 | Fixed |
| Tumor size (small vs large) | 8 | 550 | 1.19 (0.47, 2.59) | 0.83 | 81 | < 0.00001 | Random |
| Tumor grade (poor vs well+moderate) | 4 | 340 | 1.61 (1.01, 2.56) | 0.05 | 48 | 0.12 | Fixed |
| Tumor stage (high vs low) | 7 | 500 | 1.18 (0.45, 3.11) | 0.74 | 83 | < 0.00001 | Random |
| LNM (yes vs no) | 5 | 336 | 2.66 (1.60, 4.43) | 0.0002 | 47 | 0.11 | Fixed |
HR: hazard ratios; OR: odds ratios; CI: confidence intervals; OS: overall survival; MA: multivariate analysis; SC: survival curve; LNM: lymph node metastasis.
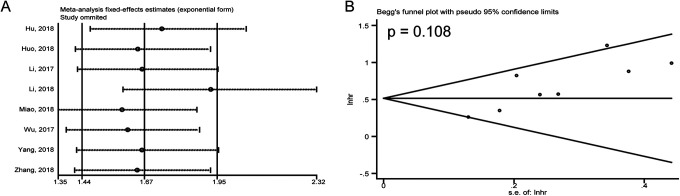
Otherwise, Begg’s test showed that there existed no significant publication bias in the meta-analysis of OS (P = 0.108) (Figure 3B), and sensitivity analysis did not detect obvious change in the combined HRs results after removing any individual study (Figure 3A).
Figure 3.
(A) Sensitivity analysis for the meta-analysis of OS; (B) Begg’s test for the meta-analysis of OS.
Association Between LncRNA NORAD Expression and Clinicopathological Parameters
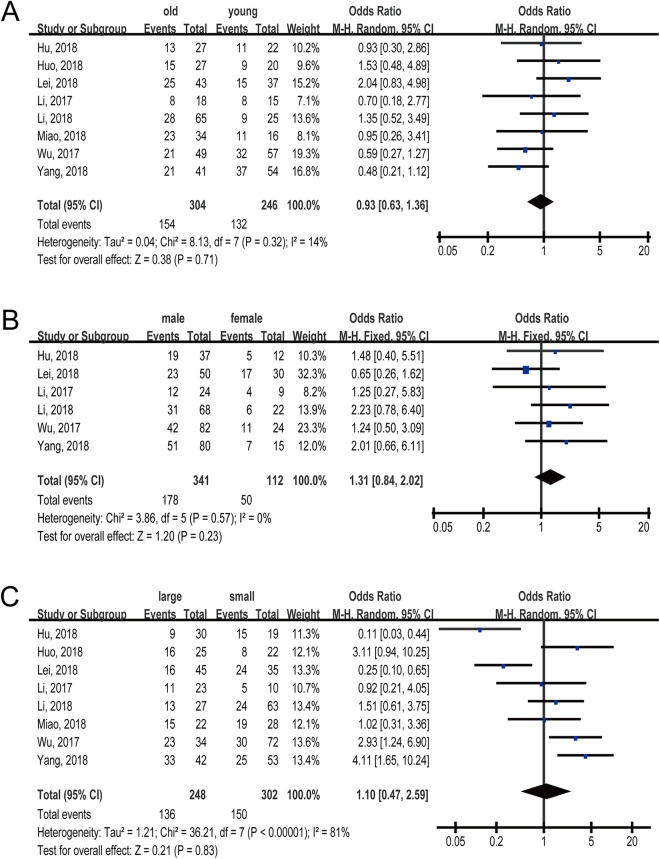
To investigate the correlation between lncRNA NORAD expression and clinicopathological parameters of cancer patients, we performed further meta-analysis for the studies that provided relative data in papers, including gender, age, tumor size, tumor grade, tumor stage, and LNM. The results demonstrated that elevated lncRNA NORAD expression was only correlated with poor tumor grade (OR = 1.61; 95% CI, 1.01-2.56; P = 0.05) (Figure 4A) and positive LNM (OR = 2.66; 95% CI, 1.60-4.43; P = 0.0002) (Figure 4C); while we failed to identify significant association between lncRNA NORAD expression level and other parameters, such as age (OR = 0.93; 95% CI, 0.63-1.36; P = 0.71) (Figure 5A), gender (OR = 1.31; 95% CI, 0.84-2.02; P = 0.23) (Figure 5B), tumor size (OR = 1.10; 95% CI, 0.47-2.59; P = 0.83) (Figure 5C), and tumor stage (OR = 1.18; 95% CI, 0.45-3.11; P = 0.74 (Figure 4B). The pooled results were presented in Table 2.
Figure 4.
Forest plots for the relationship between NORAD expression and clinicopathological parameters: (A) tumor grade; (B) tumor stage; (C) lymph node metastasis.
Figure 5.
Forest plots for the relationship between lncRNA NORAD expression and clinicopathological parameters: (A) age; (B) gender; (C) tumor size.
Discussion
LncRNAs have been confirmed to be involved in many human diseases, including malignant cancers.29,30 Many studies have reported that several lncRNAs could be used as prognostic biomarkers in human cancers, such as MALAT1, GAS5, XIST, and BLACAT1.31-34 LncRNA NORAD is a novel identified lncRNA, located on Chr20q11.23. Lee et al first discovered that lncRNA NORAD could maintain genomic stability in human cells.16 Moreover, lncRNA NORAD is highly conserved and abundantly expressed; and it usually functions as a defender of the genome mainly by regulating the activity of RNA binding proteins, such as PUM1 and PUM2.15 In recent years, several studies have indicated that high lncRNA NORAD expression was correlated with worse prognosis in certain types of cancer, such as bladder cancer, esophageal squamous cell carcinoma, colorectal cancer, and breast cancer.19,26,35,36 However, there is still no comprehensive meta-analysis conducted to investigate the prognostic value of lncRNA NORAD in various human cancers.
In the present study, we found that elevated lncRNA NORAD expression was significantly correlated with reduced OS (HR = 1.67; 95% CI, 1.44-1.95; P < 0.00001) and poor tumor stage (OR = 1.61; 95% CI, 1.01-2.56; P = 0.05). Furthermore, patients with high lncRNA NORAD expression in cancer tissues were more likely to develop LNM (OR = 2.66; 95% CI, 1.60-4.43; P = 0.0002). All these results demonstrated that lncRNA NORAD could be used as a novel predictor of worse clinical outcomes for cancer patients. However, the molecular mechanisms underlying the promotive effect of increased lncRNA NORAD expression level on progression of human cancers remains unclear.
Lots of cancer researchers have taken great efforts to explore the functional mechanisms of lncRNA NORAD on tumorigenesis and progression in various malignant tumors. It was known that lncRNA NORAD could sequester PUMILIO proteins, which provided a new understanding of the relationship between lncRNAs and chromosomal stability. PUM proteins are the main functional targets of lncRNA NORAD, recent studies revealed that PUM proteins might be involved in several classic pathways in tumorigenesis, such as MAPK and p53 signal pathways, meanwhile PUM2 also could regulate the activity of E2F3.37 Moreover, Kawasaki et al reported that lncRNA NORAD could activate the TGF-β signal pathway and regulate the epithelial-to-mesenchymal transition (EMT) induced by TGF-β in human lung cancer cells (A549).38 Li et al also suggested that lncRNA NORAD overexpression enhanced hypoxia-induced EMT and resulted in higher invasiveness of pancreatic cancer cell lines, possibly through the mechanism that lncRNA NORAD served as a competing endogenous RNA (ceRNA) to modulate the RhoA expression by sponging with miR-125a-3p.18 SIP1 was a transcriptional factor, which collaborated with the TGF-β signal pathway through interaction with Smad factors;39 Huo et al revealed that lncRNA NORAD promoted the proliferation and invasion of cervical cancer cells by upregulating SIP1 expression via lncRNA NORAD/miR-590-3p/SIP1 axis.23 MiR-202-5p functioned as an important tumor-suppressing miRNA by suppressing the TGF-β signal pathway, it has been reported that lncRNA NORAD could enhance the TGF-β signal pathway by sponging miR-202-5p to promote the progression of hepatocellular carcinoma and colorectal cancer.17,27 All in all, these studies indicated that lncRNA NORAD might be used as a promising target for the treatment of human cancers.
Our study was the first qualified meta-analysis to evaluate the prognostic value of lncRNA NORAD in human cancers, suggesting that lncRNA NORAD could become a promising predictor of lymph node metastasis and prognosis for solid tumors. Nevertheless, several limitations were still noted. Firstly, only 9 studies were finally included in our meta-analysis and their sample sizes were relatively small, these might influence the firmness of the combined results. Secondly, all the studies were conducted in China, the obtained results were not probably representative for non-China patients. Thirdly, the cutoff value in each study was not consistent for the differentiation of the high and low lncRNA NORAD expression group, thus possibly limiting the use of the conclusions in clinical practice. Finally, the HRs and their 95% CIs of several studies in our meta-analysis were calculated from the survival curves, the results might be influenced by subjective factors of the operators. Notwithstanding the foregoing, our study results demonstrated that lncRNA NORAD had good prognostic value in human cancer.
Conclusion
Taken together, this comprehensive meta-analysis clearly indicated that cancer patients with elevated lncRNA NORAD expression might have worse clinical outcomes in future. And lncRNA NORAD could be a valuable biomarker to predict the prognosis and LNM in various types of cancer. However, in terms of the several limitations mentioned before, additional studies with larger sample size, non-Chinese participants, and better design are needed to confirm our results and highlight the prognostic value of lncRNA NORAD.
Supplemental Material
Supplemental Material, S1_Table._PRISMA_checklist for LncRNA NORAD as a Novel Predictor of Lymph Node Metastasis and Prognosis in Solid Tumors: A Systematic Review and Meta-Analysis by Tao Ye and Zhangqun Ye in Technology in Cancer Research & Treatment
Supplemental Material, S2_Table._AMSTAR_checklist for LncRNA NORAD as a Novel Predictor of Lymph Node Metastasis and Prognosis in Solid Tumors: A Systematic Review and Meta-Analysis by Tao Ye and Zhangqun Ye in Technology in Cancer Research & Treatment
Acknowledgments
Authors thanks very much for the efforts and work Dr. Musa Male have done.
Abbreviations
- NORAD
Non-coding RNA-activated by DNA damage
- OS
overall survival
- LNM
lymph node metastasis
- DM
distant metastasis
- HR
hazard ratio
- CI
confidence interval
- OR
odds ratio
- NOS
Newcastle-Ottawa scale
- PC
pancreatic cancer
- BC
bladder cancer
- GC
gastric cancer
- ESCC
esophageal squamous cell carcinoma
- HCC
hepatocellular carcinoma
- CRC
colorectal cancer.
Authors’ Note: All relevant data are within the paper and its Supporting Information files. This is a meta-analysis based on previously published studies.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Tao Ye  https://orcid.org/0000-0003-4492-8399
https://orcid.org/0000-0003-4492-8399
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 3. Mirzaei HR, Sahebkar A, Salehi R, et al. Boron neutron capture therapy: moving toward targeted cancer therapy. J Cancer Res Ther. 2016;12(2):520–525. [DOI] [PubMed] [Google Scholar]
- 4. Mirzaei HR, Mirzaei H, Lee SY, Hadjati J, Till BG. Prospects for chimeric antigen receptor (CAR) γδ T cells: a potential game changer for adoptive T cell cancer immunotherapy. Cancer Lett. 2016;380(2):413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keshavarzi M, Darijani M, Momeni F, et al. Molecular imaging and oral cancer diagnosis and therapy. J Cell Biochem. 2017;118(10):3055–3060. [DOI] [PubMed] [Google Scholar]
- 6. GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1736–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research. Oncogene. 2012;31(43):4577–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esteller M, Pandolfi PP. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 2017;7(4):359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234(10):17064–17099. [DOI] [PubMed] [Google Scholar]
- 11. Naeli P, Pourhanifeh MH, Karimzadeh MR, et al. Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol. 2020;145:102854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koch L. Functional genomics: screening for lncRNA function. Nat Rev Genet. 2017;18(2):70. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Pitchiaya S, Cieślik M, et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet. 2018;50(6):814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–720. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ventura A. NORAD: defender of the genome. Trends Genet. 2016;32(7):390–392. [DOI] [PubMed] [Google Scholar]
- 16. Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164(1-2):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J, Li XY, Hu P, Ding YS. LncRNA NORAD contributes to colorectal cancer progression by inhibition of miR-202-5p. Oncol Res. 2018;26(9):1411–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Wang X, Wen C, et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16(1):169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Q, Li C, Chen J, et al. High expression of long noncoding RNA NORAD indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Urol Oncol. 2018;36(6):310.e15–310.e22. [DOI] [PubMed] [Google Scholar]
- 20. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–341. [DOI] [PubMed] [Google Scholar]
- 21. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. [DOI] [PubMed] [Google Scholar]
- 22. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huo H, Tian J, Wang R, Li Y, Qu C, Wang N. Long non-coding RNA NORAD upregulate SIP1 expression to promote cell proliferation and invasion in cervical cancer. Biomed Pharmacother. 2018;106:1454–1460. [DOI] [PubMed] [Google Scholar]
- 24. Lei Y, Wang YH, Wang XF, Bai J. LINC00657 promotes the development of colon cancer by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22(19):6315–6323. [DOI] [PubMed] [Google Scholar]
- 25. Miao Z, Guo X, Tian L. The long noncoding RNA NORAD promotes the growth of gastric cancer cells by sponging miR-608. Gene. 2018;687:116–124. [DOI] [PubMed] [Google Scholar]
- 26. Wu X, Lim ZF, Li Z, et al. NORAD expression is associated with adverse prognosis in esophageal squamous cell carcinoma. Oncol Res Treat. 2017;40(6):370–374. [DOI] [PubMed] [Google Scholar]
- 27. Yang X, Cai JB, Peng R, et al. The long noncoding RNA NORAD enhances the TGF-β pathway to promote hepatocellular carcinoma progression by targeting miR-202-5p. J Cell Physiol. 2018;234(7):12051–12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu B, Cai H, Zheng R, Yang S, Zhou Z, Tu J. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int J Biochem Cell Biol. 2017;92:34–42. [DOI] [PubMed] [Google Scholar]
- 29. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. [DOI] [PubMed] [Google Scholar]
- 30. Vafadar A, Shabaninejad Z, Movahedpour A, et al. Long non-coding RNAs as epigenetic regulators in cancer. Curr Pharm Des. 2019;25(33):3563–3577. [DOI] [PubMed] [Google Scholar]
- 31. Wei Y, Niu B. Role of MALAT1 as a prognostic factor for survival in various cancers: a systematic review of the literature with meta-analysis. Dis Markers. 2015;2015:164635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song W, Wang K, Zhang RJ, Dai QX, Zou SB. Long noncoding RNA GAS5 can predict metastasis and poor prognosis: a meta-analysis. Minerva Med. 2016;107(1):70–76. [PubMed] [Google Scholar]
- 33. Liu X, Ming X, Jing W, et al. Long non-coding RNA XIST predicts worse prognosis in digestive system tumors: a systemic review and meta-analysis. Biosci Rep. 2018;38(3):BSR20180169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu H, Liu H, Yang X, et al. LncRNA BLACAT1 may serve as a prognostic predictor in cancer: evidence from a meta-analysis. Biomed Res Int. 2019;2019:1275491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L, Du L, Duan W, Yan S, Xie Y, Wang C. Overexpression of long noncoding RNA NORAD in colorectal cancer associates with tumor progression. Onco Targets Ther. 2018;11:6757–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H, Li J, Koirala P, et al. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016;7(15):20584–20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miles WO, Tschöp K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26(4):356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kawasaki N, Miwa T, Hokari S, et al. Long noncoding RNA NORAD regulates transforming growth factor-β signaling and epithelial-to-mesenchymal transition-like phenotype. Cancer Sci. 2018;109(7):2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyoshi A, Kitajima Y, Sumi K, et al. Snail and SIP1 increase cancer invasion by upregulating MMP family in hepatocellular carcinoma cells. Br J Cancer. 2004;90(6):1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, S1_Table._PRISMA_checklist for LncRNA NORAD as a Novel Predictor of Lymph Node Metastasis and Prognosis in Solid Tumors: A Systematic Review and Meta-Analysis by Tao Ye and Zhangqun Ye in Technology in Cancer Research & Treatment
Supplemental Material, S2_Table._AMSTAR_checklist for LncRNA NORAD as a Novel Predictor of Lymph Node Metastasis and Prognosis in Solid Tumors: A Systematic Review and Meta-Analysis by Tao Ye and Zhangqun Ye in Technology in Cancer Research & Treatment