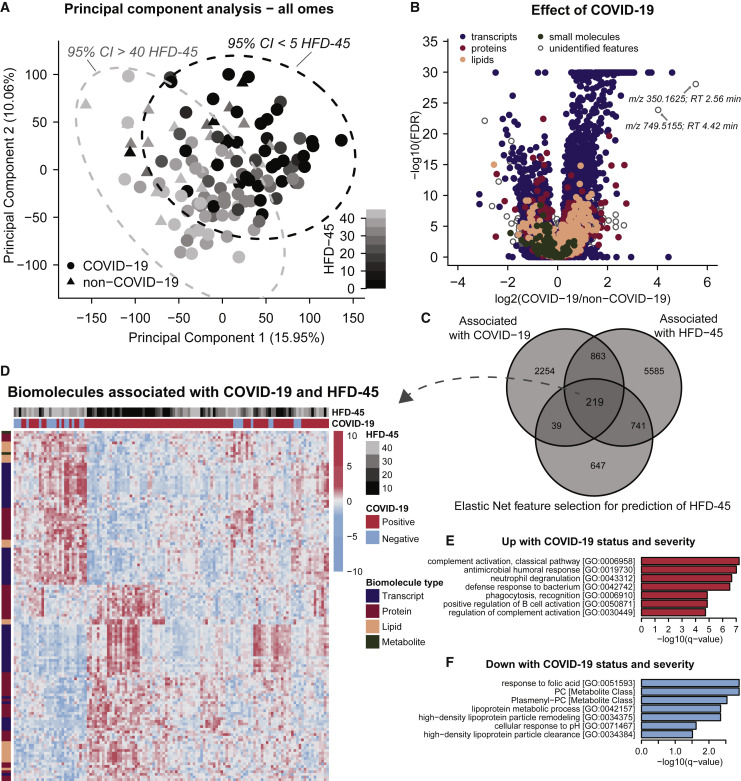
Figure 2.
Multi-omics Analysis Reveals Strong Molecular Signatures Associated with COVID-19 Status and Severity
(A) PCA using quantitative values from all omics data (leukocyte transcripts, and plasma proteins, lipids, and small molecules, log2 transformed and centered around 0, for n = 125 patient samples, also see STAR Methods) shows that principal components 1 and 2 capture 16% and 10%, respectively, of the variance between patient samples. Plotting samples by these two components show a linear trend with hospital-free days at 45 days (HFD-45).
(B) Associations of biomolecules with COVID-19 status were determined using differential expression analysis (EB-seq) for transcripts and linear regression log-likelihood tests for plasma biomolecules (see Table S1). The adjusted p values (1 - posterior probability or Benjamini Hochberg-adjusted p values, respectively) are plotted relative to the log2 fold-change of mean values between COVID and non-COVID samples. In total, 2,537 leukocyte transcripts, 146 plasma proteins, 168 plasma lipids, and 13 plasma metabolites had adjusted p values < 0.05.
(C) Associations between biomolecules and HFD-45 was estimated using a univariate linear regression (HFD-45 ~ biomolecule abundance + age + sex) resulting in 7,408 biomolecules significantly associated with HFD-45, see Table S1. A multivariate linear regression with elastic net penalty was applied to each omics dataset separately to further refine features of interest, and resulted in 946 features that were retained as coefficients predictive for HFD-45, also see Table S1. In total, 219 features were determined as most important for distinguishing COVID status and severity.
(D) Abundance of the 219 features were visualized via a heatmap (Z scored by row) and clustered with hierarchical clustering.
(E and F) Features that were elevated (E) or reduced (F) with COVID-19 status and severity were used for GO term and molecular class enrichment analysis (see Table S2).