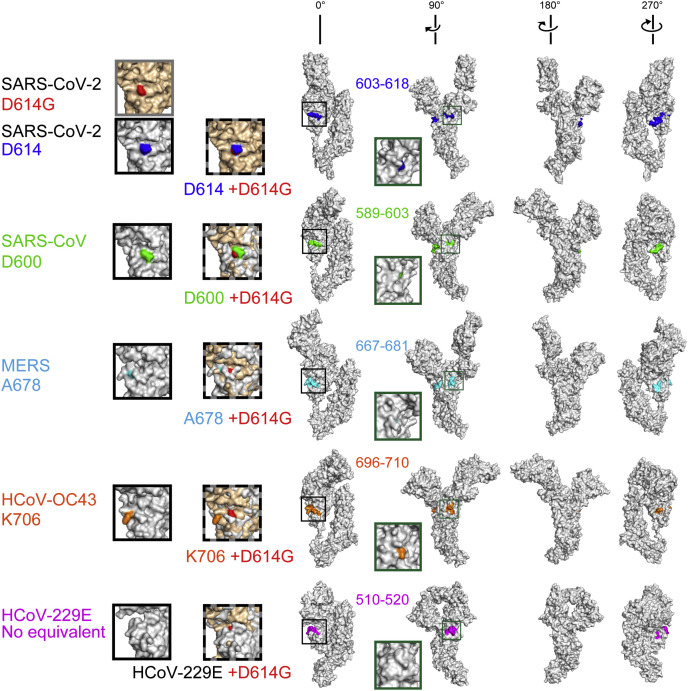
Fig. 2.
Comparison of D614 and equivalent residues across coronaviruses.
The entire surface views of the available spike crystal structures of SARS-CoV-2, SARS-CoV, MERS, HCoV-OC
43 and HCoV-229E. See also the grey bar in Fig. 1b and 1c. The SARS-CoV-2 S protein with D614G conversion is indicated in beige, and the other spikes are in light grey; the aligned structures are shown in mosaic colours. Homologous regions surrounding D614 of SARS-CoV-2 are highlighted with distinct colours. A box with grey solid lines shows a close-up view of D614G conversion in SARS-CoV-2. Boxes with black solid lines show the equivalent residues in other CoVs. Boxes with black-grey dotted lines show the structural alignment of equivalent regions between SARS-CoV-2:D614G and the other respective CoVs. HCoV-229E lacks the residue equivalent to D614 of SARS-CoV-2. Boxes in green indicate the close-up view of the equivalent residues rotated by 90°; not overlaid. The data for HCoV-HKU1 and HCoV–NL63 were not found in the database. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)