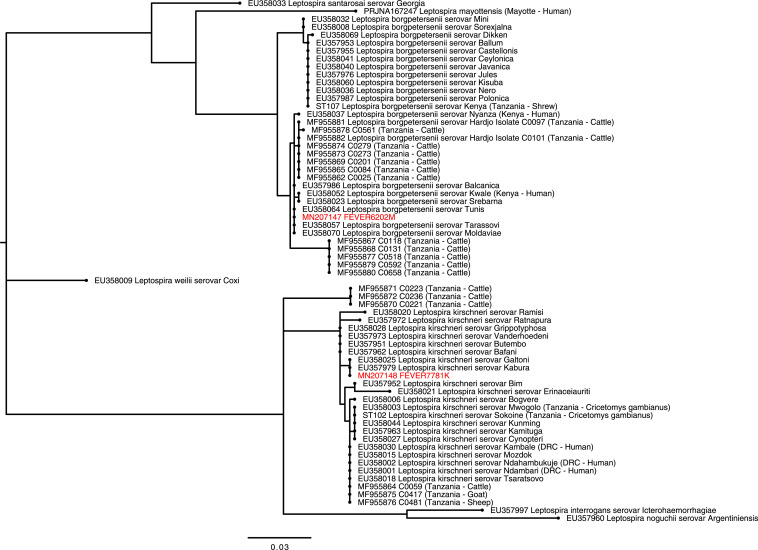
Figure 1.
Phylogenetic tree showing the relationship between Leptospira secY gene (434-bp fragment) derived from human infections in Tanzania with sequence from reference serovars8 and previously published sequences from human and animal infection in Tanzania4,27 The phylogenetic tree was constructed using the maximum likelihood method using the Tamura 3-parameter (T92) nucleotide substitution model with a discrete gamma distribution.44 The tree with the highest log likelihood is shown. Sequences from our study are labeled with unique identifiers (FEVERXXXX) and GenBank accession numbers and highlighted in red. Sequences generated from reference Leptospira serovars and from other studies in Tanzania are shown and labeled with GenBank accession numbers and ST types, respectively.45 Expanded clades show reference serovars most closely related to the study genotypes. Clades of more distantly related species are collapsed and labeled with species names only. Country locations and host are shown in parenthesis for East African studies. DRC = Democratic Republic of Congo; sv = serovar; ST = sequence types. This figure appears in color at www.ajtmh.org.