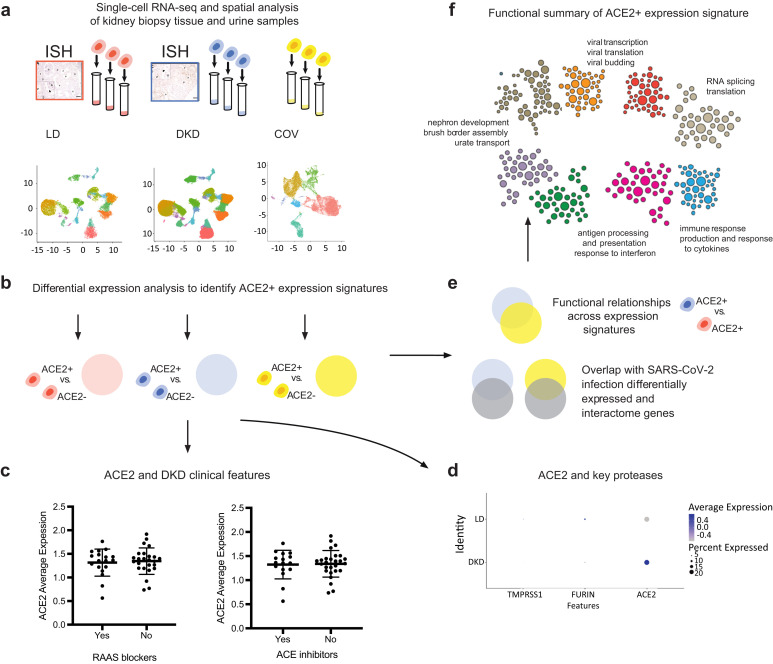
Figure 1.
Study overview. To understand how the kidney may be affected by coronavirus disease 2019 (COVID-19) we performed spatial, systems, and clinical association analyses of angiotensin-converting enzyme 2 (ACE2) and other severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) host factors in kidney biopsies from living donors (LD) and patients with diabetic kidney disease (DKD) and kidney cells isolated from the urine of hospitalized COVID-19 patients (COV). (a) Biopsy samples from DKD and LD were processed for in situ hybridization (ISH) and single-cell RNA sequencing (scRNAseq) profiling. scRNAseq of LD, DKD, and urine cell pellets from COV samples were analyzed to determine cell type expression specificity of ACE2 in healthy and disease states. (b) For each scRNAseq dataset, ACE2+ differential expression signatures were identified. (c) Association of ACE2 expression levels in DKD with clinical characteristics were evaluated, including exposure to renin-angiotensin-aldosterone system (RAAS) blockers and ACE inhibitors. (d) Expression of ACE2 and key proteases between LD and DKD proximal tubule epithelial cells (PTECs) were compared. (e) ACE2 expression signatures across datasets identified aspects induced in PTECs expressing DKD samples compared to LD. These gene sets significantly overlapped those reported to be affected by direct SARS-CoV-2 infection. (f) The biological processes in ACE2+ expression signatures were characterized by projecting these signature genes onto PTEC-specific functional networks at HumanBase (https://hb.flatironinstitute.org/covid-kidney). These networks represent genes and their interactions in biological processes and pathways active in PTECs.