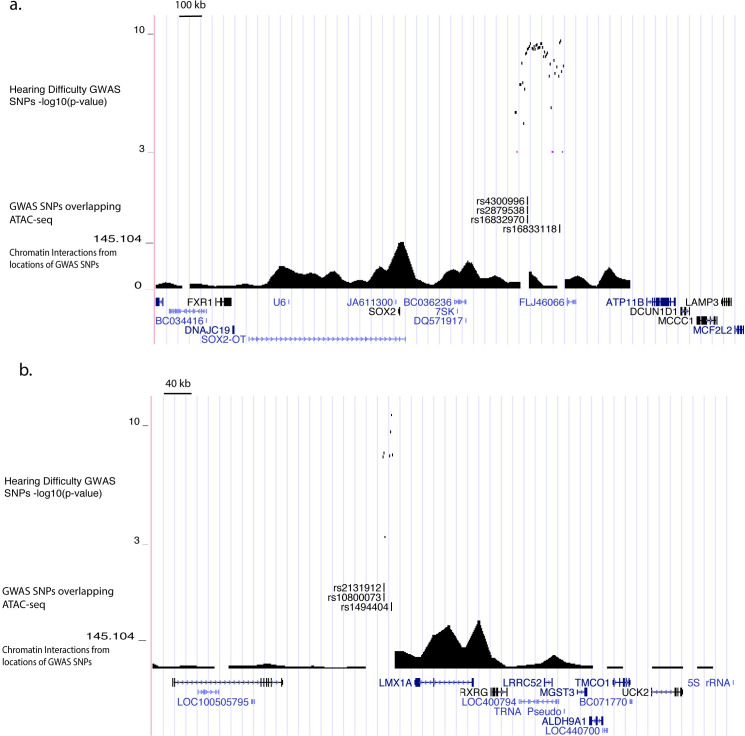
Fig 4. Epigenomic fine-mapping predicts distal target genes for hearing difficulty risk loci.
Genetic associations and epigenomic annotations at chr3q26.3 (a) and chr1q23.3 (b). From top to bottom, genome browser tracks indicate: -log10(p-values) for association with hearing difficulty; fine-mapped SNPs in strong LD with an LD-independent lead SNP and located <500bp from a region homologous to a cochlear open chromatin region based on ATAC-seq; -log10(p-values) for chromatin interactions between the locations of the fine-mapped SNPs and distal regions, based on the minimum chromatin interaction p-value in each 40kb region from Hi-C of 20 non-cochlear human tissues and cell types[44]; locations of UCSC knownGene gene models.