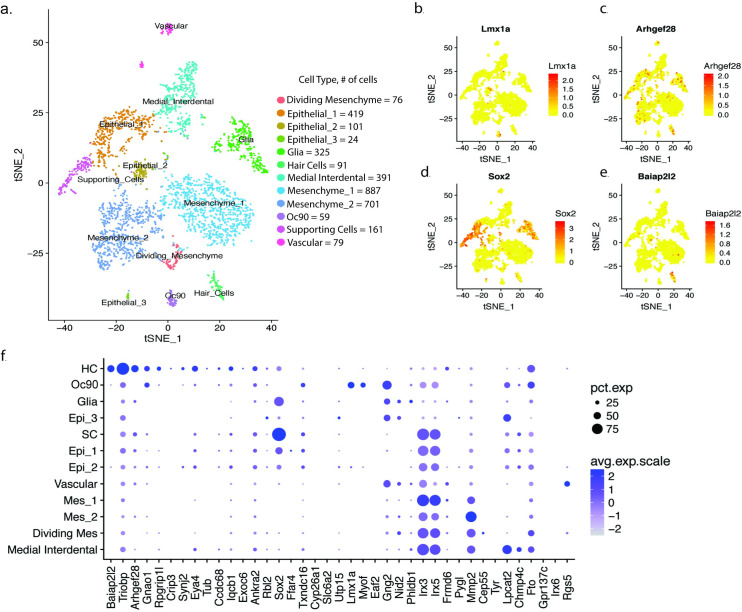
Fig 5. Single-cell RNA-seq of mouse cochlea reveals cell type-specific expression patterns of hearing difficulty risk genes.
a. t-distributed stochastic neighbor embedding (t-SNE) plot of 3,411 cells in the postnatal day 2 mouse cochlea colored by Louvain modularity clusters corresponding to 12 cell types. b-e. t-SNE plots colored by the expression of selected hearing difficulty risk genes expressed selectively in cochlear cell types: LMX1A in a subset of epithelial Oc90 cells (b); ARHGEF28 in hair cells (c), SOX2 in supporting cells (d), and BAIAP2L2 in hair cells (e). f. Dot plot showing the average expression and percent of cells with non-zero counts for each cochlea-expressed risk gene in each of the 12 cochlear cell types.