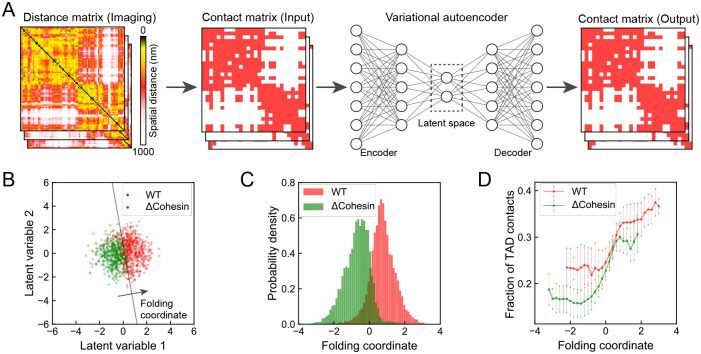
Fig 2. Chromatin folding coordinate derived using deep learning to differentiate chromatin organization in WT and cohesin-depleted cells.
(A) Illustration of the variational autoencoder for data processing and low-dimensional embedding. Single-cell chromatin images were first binarized into contact matrices that can be fed into VAE as inputs. The encoder network further projects the high dimensional contacts into a small set of latent variables that best preserve key features of the original data. The decoder network then defines the reconstruction from latent variables to contact matrices. (B) Scatter plot for WT and cohesin-depleted cells in the two-dimensional space of latent variables learned from VAE. The black line represents the decision boundary and the folding coordinate is defined as the distance from the boundary. To avoid overplotting, only 5% of randomly sampled data are shown. For the full dataset, if all the points that fall to the lower left of the boundary were all assigned as cohesin-depleted cells and those on the upper right as WT cells, the misclassification rate is 12.8%. (C) Probability distributions of the folding coordinate for chromatin structures from WT and cohesin-depleted cells. (D) Correlation between the folding coordinate and the fraction of contacts formed within the WT TADs. Error bars represent one standard deviation of uncertainty.